diff options
Diffstat (limited to 'python-ipycytoscape.spec')
| -rw-r--r-- | python-ipycytoscape.spec | 668 |
1 files changed, 668 insertions, 0 deletions
diff --git a/python-ipycytoscape.spec b/python-ipycytoscape.spec new file mode 100644 index 0000000..1c91e1c --- /dev/null +++ b/python-ipycytoscape.spec @@ -0,0 +1,668 @@ +%global _empty_manifest_terminate_build 0 +Name: python-ipycytoscape +Version: 1.3.3 +Release: 1 +Summary: A Cytoscape widget for Jupyter +License: BSD +URL: https://github.com/cytoscape/ipycytoscape +Source0: https://mirrors.nju.edu.cn/pypi/web/packages/21/4b/dca529aa566ce225107580c6c8625c7dc5ecb1532f7d73259e2888d2187a/ipycytoscape-1.3.3.tar.gz +BuildArch: noarch + +Requires: python3-ipywidgets +Requires: python3-spectate +Requires: python3-sphinx +Requires: python3-sphinx-rtd-theme +Requires: python3-sphinx-autobuild +Requires: python3-jupyter-sphinx +Requires: python3-sphinx-copybutton +Requires: python3-nbsphinx +Requires: python3-nbsphinx-link +Requires: python3-networkx +Requires: python3-pandas +Requires: python3-pandas +Requires: python3-py2neo +Requires: python3-monotonic +Requires: python3-py2neo +Requires: python3-monotonic +Requires: python3-pytest +Requires: python3-pytest-cov +Requires: python3-nbval +Requires: python3-pandas +Requires: python3-nbclassic +Requires: python3-networkx +Requires: python3-pre-commit + +%description +# ipycytoscape + +[](https://github.com/cytoscape/ipycytoscape/actions/workflows/test.yml) [](https://ipycytoscape.readthedocs.io/en/latest/?badge=master) [](https://stackoverflow.com/questions/tagged/ipycytoscape) [](https://gitter.im/QuantStack/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) + +A widget enabling interactive graph visualization with [cytoscape.js](https://js.cytoscape.org/) in JupyterLab and the Jupyter notebook. + +Try it out using binder: [](https://mybinder.org/v2/gh/cytoscape/ipycytoscape/HEAD?filepath=examples) or install and try out the [examples](examples). + +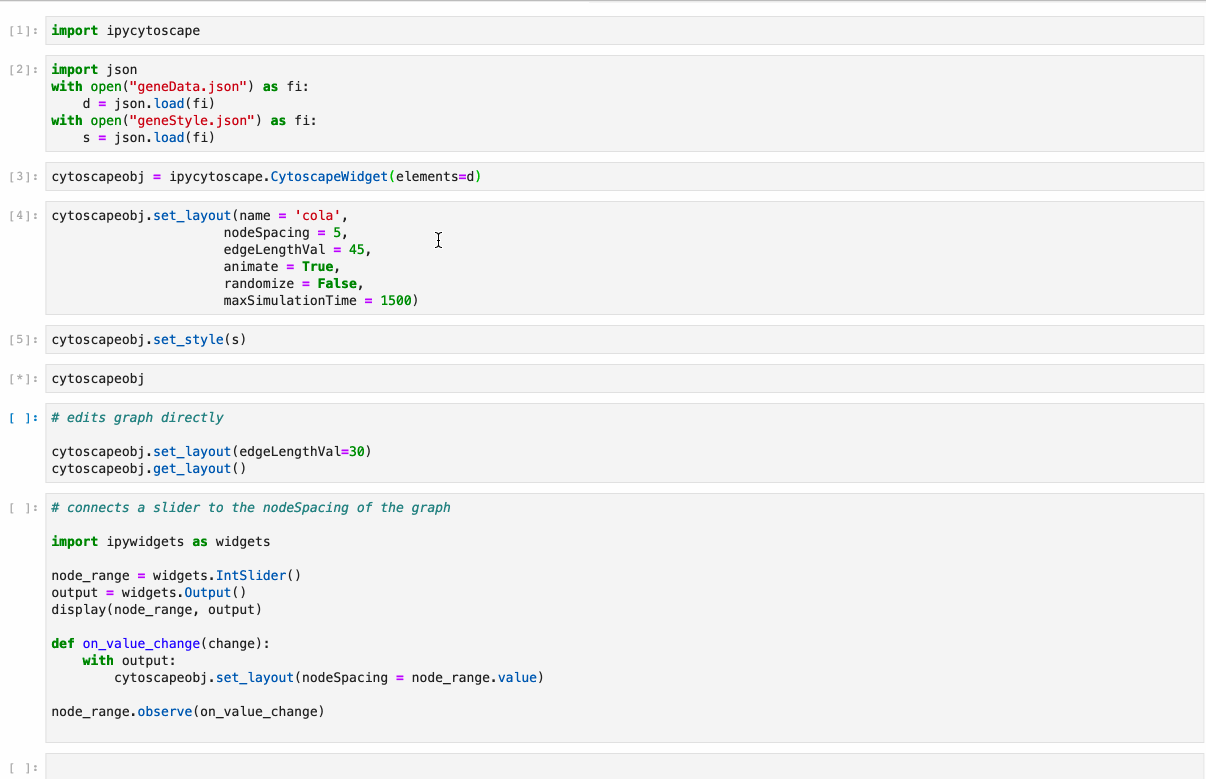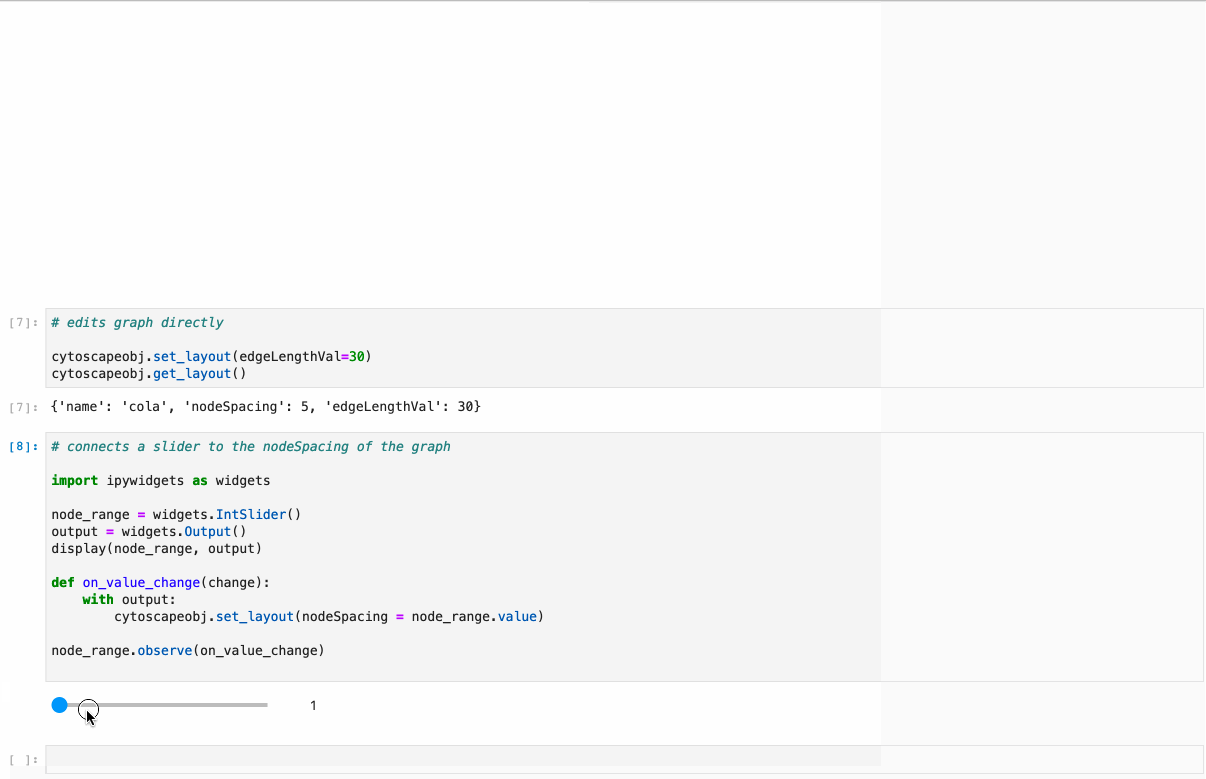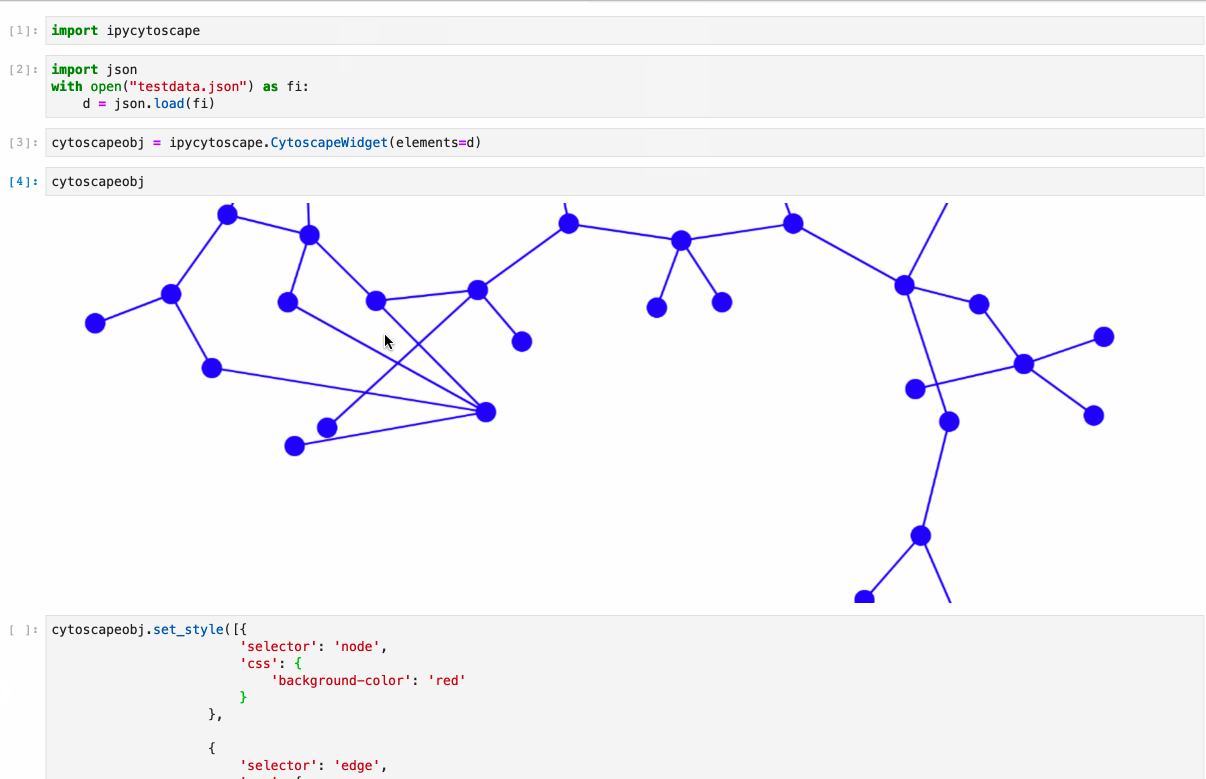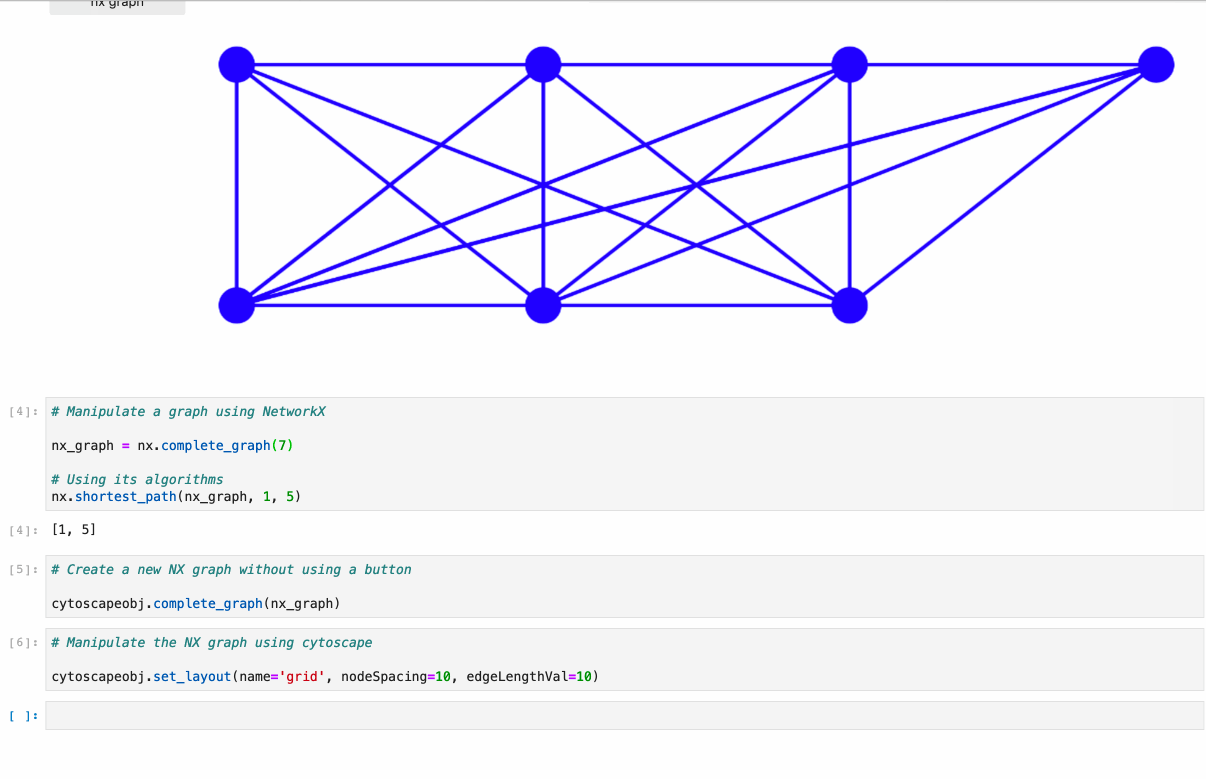 + +#### Supports: + +* Conversion from NetworkX see [example1](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Test%20NetworkX%20methods.ipynb), [example2](https://github.com/cytoscape/ipycytoscape/blob/master/examples/NetworkX%20Example.ipynb) +* Conversion from Pandas DataFrame see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/pandas.ipynb) +* Conversion from neo4j see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Neo4j_Example.ipynb) + +## Installation + +With `mamba`: + +``` +mamba install -c conda-forge ipycytoscape +``` + +With `conda`: + +``` +conda install -c conda-forge ipycytoscape +``` + +With `pip`: + +```bash +pip install ipycytoscape +``` + +### Pandas installation + +You can install the Pandas dependencies for `ipycytoscape` with pip: + +``` +pip install pandas +``` + +Or conda-forge: + +``` +mamba install pandas +``` + +### Neo4j installation + +You can install the neo4j dependencies for `ipycytoscape` with pip: + +``` +pip install -e ".[neo4j]" +``` + +Or conda-forge: +``` +mamba install py2neo neotime +``` + +#### For jupyterlab 1.x or 2.x: + +If you are using JupyterLab 1.x or 2.x then you will also need to install `nodejs` and the `jupyterlab-manager` extension. You can do this like so: + +```bash +# installing nodejs +conda install -c conda-forge nodejs + + +# install jupyterlab-manager extension +jupyter labextension install @jupyter-widgets/jupyterlab-manager@2.0 --no-build + +# if you have previously installed the manager you still to run jupyter lab build +jupyter lab build +``` + +### For Jupyter Notebook 5.2 and earlier + +You may also need to manually enable the nbextension: +```bash +jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscape +``` + +## For a development installation: +**(requires npm)** + +While not required, we recommend creating a conda environment to work in: +```bash +conda create -n ipycytoscape -c conda-forge jupyterlab nodejs +conda activate ipycytoscape + +# clone repo +git clone https://github.com/cytoscape/ipycytoscape.git +cd ipycytoscape +``` + +### Install python package for development + +This will `run npm install` and `npm run build`. +This command will also install the test suite and the [docs](https://ipycytoscape.readthedocs.io/en/latest/) locally: + +``` +pip install jupyter_packaging==0.7.9 +pip install -e ".[test, docs]" + +jupyter labextension develop . --overwrite +``` + +optionally install the pre-commit hooks with: + +```bash +pre-commit install +``` + + +Or for classic notebook, you can run: + +``` +jupyter nbextension install --sys-prefix --symlink --overwrite --py ipycytoscape +jupyter nbextension enable --sys-prefix --py ipycytoscape +``` + +Note that the `--symlink` flag doesn't work on Windows, so you will here have to run +the `install` command every time that you rebuild your extension. For certain installations +you might also need another flag instead of `--sys-prefix`, but we won't cover the meaning +of those flags here. + +### How to see your changes + +#### Typescript: +To continuously monitor the project for changes and automatically trigger a rebuild, start watching the ipycytoscape code: +```bash +npm run watch +``` +And in a separate terminal start JupyterLab normally: +```bash +jupyter lab +``` +once the webpack rebuild finishes refresh the JupyterLab page to have your changes take effect. + +#### Python: +If you make a change to the python code then you need to restart the notebook kernel to have it take effect. + +### How to run tests locally + +Install necessary dependencies with pip: + +``` +pip install -e ".[test]" +``` + +Or with conda/mamba: + +``` +mamba -c conda-forge install networkx pandas nbval pytest +``` + +#### And to run it: + +``` +pytest +``` + +### How to build the docs + +`cd docs` + +Install dependencies: + +`conda env update --file doc_environment.yml` + +And build them: + +`make html` + +## Acknowledgements + +The ipycytoscape project was started by [Mariana Meireles](https://github.com/marimeireles) at [QuantStack](https://quantstack.net). This initial development was funded as part of the [PLASMA](https://plasmabio.org) project, which is led by Claire Vandiedonck, Pierre Poulain, and Sandrine Caburet. + +## License + +We use a shared copyright model that enables all contributors to maintain the +copyright on their contributions. + +This software is licensed under the BSD-3-Clause license. See the +[LICENSE](LICENSE) file for details. + + + + +%package -n python3-ipycytoscape +Summary: A Cytoscape widget for Jupyter +Provides: python-ipycytoscape +BuildRequires: python3-devel +BuildRequires: python3-setuptools +BuildRequires: python3-pip +%description -n python3-ipycytoscape +# ipycytoscape + +[](https://github.com/cytoscape/ipycytoscape/actions/workflows/test.yml) [](https://ipycytoscape.readthedocs.io/en/latest/?badge=master) [](https://stackoverflow.com/questions/tagged/ipycytoscape) [](https://gitter.im/QuantStack/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) + +A widget enabling interactive graph visualization with [cytoscape.js](https://js.cytoscape.org/) in JupyterLab and the Jupyter notebook. + +Try it out using binder: [](https://mybinder.org/v2/gh/cytoscape/ipycytoscape/HEAD?filepath=examples) or install and try out the [examples](examples). + + + +#### Supports: + +* Conversion from NetworkX see [example1](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Test%20NetworkX%20methods.ipynb), [example2](https://github.com/cytoscape/ipycytoscape/blob/master/examples/NetworkX%20Example.ipynb) +* Conversion from Pandas DataFrame see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/pandas.ipynb) +* Conversion from neo4j see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Neo4j_Example.ipynb) + +## Installation + +With `mamba`: + +``` +mamba install -c conda-forge ipycytoscape +``` + +With `conda`: + +``` +conda install -c conda-forge ipycytoscape +``` + +With `pip`: + +```bash +pip install ipycytoscape +``` + +### Pandas installation + +You can install the Pandas dependencies for `ipycytoscape` with pip: + +``` +pip install pandas +``` + +Or conda-forge: + +``` +mamba install pandas +``` + +### Neo4j installation + +You can install the neo4j dependencies for `ipycytoscape` with pip: + +``` +pip install -e ".[neo4j]" +``` + +Or conda-forge: +``` +mamba install py2neo neotime +``` + +#### For jupyterlab 1.x or 2.x: + +If you are using JupyterLab 1.x or 2.x then you will also need to install `nodejs` and the `jupyterlab-manager` extension. You can do this like so: + +```bash +# installing nodejs +conda install -c conda-forge nodejs + + +# install jupyterlab-manager extension +jupyter labextension install @jupyter-widgets/jupyterlab-manager@2.0 --no-build + +# if you have previously installed the manager you still to run jupyter lab build +jupyter lab build +``` + +### For Jupyter Notebook 5.2 and earlier + +You may also need to manually enable the nbextension: +```bash +jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscape +``` + +## For a development installation: +**(requires npm)** + +While not required, we recommend creating a conda environment to work in: +```bash +conda create -n ipycytoscape -c conda-forge jupyterlab nodejs +conda activate ipycytoscape + +# clone repo +git clone https://github.com/cytoscape/ipycytoscape.git +cd ipycytoscape +``` + +### Install python package for development + +This will `run npm install` and `npm run build`. +This command will also install the test suite and the [docs](https://ipycytoscape.readthedocs.io/en/latest/) locally: + +``` +pip install jupyter_packaging==0.7.9 +pip install -e ".[test, docs]" + +jupyter labextension develop . --overwrite +``` + +optionally install the pre-commit hooks with: + +```bash +pre-commit install +``` + + +Or for classic notebook, you can run: + +``` +jupyter nbextension install --sys-prefix --symlink --overwrite --py ipycytoscape +jupyter nbextension enable --sys-prefix --py ipycytoscape +``` + +Note that the `--symlink` flag doesn't work on Windows, so you will here have to run +the `install` command every time that you rebuild your extension. For certain installations +you might also need another flag instead of `--sys-prefix`, but we won't cover the meaning +of those flags here. + +### How to see your changes + +#### Typescript: +To continuously monitor the project for changes and automatically trigger a rebuild, start watching the ipycytoscape code: +```bash +npm run watch +``` +And in a separate terminal start JupyterLab normally: +```bash +jupyter lab +``` +once the webpack rebuild finishes refresh the JupyterLab page to have your changes take effect. + +#### Python: +If you make a change to the python code then you need to restart the notebook kernel to have it take effect. + +### How to run tests locally + +Install necessary dependencies with pip: + +``` +pip install -e ".[test]" +``` + +Or with conda/mamba: + +``` +mamba -c conda-forge install networkx pandas nbval pytest +``` + +#### And to run it: + +``` +pytest +``` + +### How to build the docs + +`cd docs` + +Install dependencies: + +`conda env update --file doc_environment.yml` + +And build them: + +`make html` + +## Acknowledgements + +The ipycytoscape project was started by [Mariana Meireles](https://github.com/marimeireles) at [QuantStack](https://quantstack.net). This initial development was funded as part of the [PLASMA](https://plasmabio.org) project, which is led by Claire Vandiedonck, Pierre Poulain, and Sandrine Caburet. + +## License + +We use a shared copyright model that enables all contributors to maintain the +copyright on their contributions. + +This software is licensed under the BSD-3-Clause license. See the +[LICENSE](LICENSE) file for details. + + + + +%package help +Summary: Development documents and examples for ipycytoscape +Provides: python3-ipycytoscape-doc +%description help +# ipycytoscape + +[](https://github.com/cytoscape/ipycytoscape/actions/workflows/test.yml) [](https://ipycytoscape.readthedocs.io/en/latest/?badge=master) [](https://stackoverflow.com/questions/tagged/ipycytoscape) [](https://gitter.im/QuantStack/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) + +A widget enabling interactive graph visualization with [cytoscape.js](https://js.cytoscape.org/) in JupyterLab and the Jupyter notebook. + +Try it out using binder: [](https://mybinder.org/v2/gh/cytoscape/ipycytoscape/HEAD?filepath=examples) or install and try out the [examples](examples). + + + +#### Supports: + +* Conversion from NetworkX see [example1](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Test%20NetworkX%20methods.ipynb), [example2](https://github.com/cytoscape/ipycytoscape/blob/master/examples/NetworkX%20Example.ipynb) +* Conversion from Pandas DataFrame see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/pandas.ipynb) +* Conversion from neo4j see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Neo4j_Example.ipynb) + +## Installation + +With `mamba`: + +``` +mamba install -c conda-forge ipycytoscape +``` + +With `conda`: + +``` +conda install -c conda-forge ipycytoscape +``` + +With `pip`: + +```bash +pip install ipycytoscape +``` + +### Pandas installation + +You can install the Pandas dependencies for `ipycytoscape` with pip: + +``` +pip install pandas +``` + +Or conda-forge: + +``` +mamba install pandas +``` + +### Neo4j installation + +You can install the neo4j dependencies for `ipycytoscape` with pip: + +``` +pip install -e ".[neo4j]" +``` + +Or conda-forge: +``` +mamba install py2neo neotime +``` + +#### For jupyterlab 1.x or 2.x: + +If you are using JupyterLab 1.x or 2.x then you will also need to install `nodejs` and the `jupyterlab-manager` extension. You can do this like so: + +```bash +# installing nodejs +conda install -c conda-forge nodejs + + +# install jupyterlab-manager extension +jupyter labextension install @jupyter-widgets/jupyterlab-manager@2.0 --no-build + +# if you have previously installed the manager you still to run jupyter lab build +jupyter lab build +``` + +### For Jupyter Notebook 5.2 and earlier + +You may also need to manually enable the nbextension: +```bash +jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscape +``` + +## For a development installation: +**(requires npm)** + +While not required, we recommend creating a conda environment to work in: +```bash +conda create -n ipycytoscape -c conda-forge jupyterlab nodejs +conda activate ipycytoscape + +# clone repo +git clone https://github.com/cytoscape/ipycytoscape.git +cd ipycytoscape +``` + +### Install python package for development + +This will `run npm install` and `npm run build`. +This command will also install the test suite and the [docs](https://ipycytoscape.readthedocs.io/en/latest/) locally: + +``` +pip install jupyter_packaging==0.7.9 +pip install -e ".[test, docs]" + +jupyter labextension develop . --overwrite +``` + +optionally install the pre-commit hooks with: + +```bash +pre-commit install +``` + + +Or for classic notebook, you can run: + +``` +jupyter nbextension install --sys-prefix --symlink --overwrite --py ipycytoscape +jupyter nbextension enable --sys-prefix --py ipycytoscape +``` + +Note that the `--symlink` flag doesn't work on Windows, so you will here have to run +the `install` command every time that you rebuild your extension. For certain installations +you might also need another flag instead of `--sys-prefix`, but we won't cover the meaning +of those flags here. + +### How to see your changes + +#### Typescript: +To continuously monitor the project for changes and automatically trigger a rebuild, start watching the ipycytoscape code: +```bash +npm run watch +``` +And in a separate terminal start JupyterLab normally: +```bash +jupyter lab +``` +once the webpack rebuild finishes refresh the JupyterLab page to have your changes take effect. + +#### Python: +If you make a change to the python code then you need to restart the notebook kernel to have it take effect. + +### How to run tests locally + +Install necessary dependencies with pip: + +``` +pip install -e ".[test]" +``` + +Or with conda/mamba: + +``` +mamba -c conda-forge install networkx pandas nbval pytest +``` + +#### And to run it: + +``` +pytest +``` + +### How to build the docs + +`cd docs` + +Install dependencies: + +`conda env update --file doc_environment.yml` + +And build them: + +`make html` + +## Acknowledgements + +The ipycytoscape project was started by [Mariana Meireles](https://github.com/marimeireles) at [QuantStack](https://quantstack.net). This initial development was funded as part of the [PLASMA](https://plasmabio.org) project, which is led by Claire Vandiedonck, Pierre Poulain, and Sandrine Caburet. + +## License + +We use a shared copyright model that enables all contributors to maintain the +copyright on their contributions. + +This software is licensed under the BSD-3-Clause license. See the +[LICENSE](LICENSE) file for details. + + + + +%prep +%autosetup -n ipycytoscape-1.3.3 + +%build +%py3_build + +%install +%py3_install +install -d -m755 %{buildroot}/%{_pkgdocdir} +if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi +if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi +if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi +if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi +pushd %{buildroot} +if [ -d usr/lib ]; then + find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/lib64 ]; then + find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/bin ]; then + find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/sbin ]; then + find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst +fi +touch doclist.lst +if [ -d usr/share/man ]; then + find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst +fi +popd +mv %{buildroot}/filelist.lst . +mv %{buildroot}/doclist.lst . + +%files -n python3-ipycytoscape -f filelist.lst +%dir %{python3_sitelib}/* + +%files help -f doclist.lst +%{_docdir}/* + +%changelog +* Fri May 05 2023 Python_Bot <Python_Bot@openeuler.org> - 1.3.3-1 +- Package Spec generated |
