diff options
| -rw-r--r-- | .gitignore | 1 | ||||
| -rw-r--r-- | python-miscnn.spec | 282 | ||||
| -rw-r--r-- | sources | 1 |
3 files changed, 284 insertions, 0 deletions
@@ -0,0 +1 @@ +/miscnn-1.4.0.tar.gz diff --git a/python-miscnn.spec b/python-miscnn.spec new file mode 100644 index 0000000..e5fdf00 --- /dev/null +++ b/python-miscnn.spec @@ -0,0 +1,282 @@ +%global _empty_manifest_terminate_build 0 +Name: python-miscnn +Version: 1.4.0 +Release: 1 +Summary: Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning +License: GPLv3 +URL: https://github.com/frankkramer-lab/MIScnn +Source0: https://mirrors.nju.edu.cn/pypi/web/packages/3e/c6/3368ea5168d440a809264a0815beb9b26516c137a1fb106c3596e0f5125e/miscnn-1.4.0.tar.gz +BuildArch: noarch + +Requires: python3-tensorflow +Requires: python3-tensorflow-addons +Requires: python3-numpy +Requires: python3-pandas +Requires: python3-tqdm +Requires: python3-nibabel +Requires: python3-matplotlib +Requires: python3-pillow +Requires: python3-batchgenerators +Requires: python3-pydicom +Requires: python3-SimpleITK +Requires: python3-scikit-image + +%description + + +[](https://www.python.org/) +[](https://travis-ci.org/github/frankkramer-lab/MIScnn) +[](https://codecov.io/gh/frankkramer-lab/miscnn) +[](https://pypi.org/project/miscnn/) +[](https://pypistats.org/packages/miscnn) +[](https://www.gnu.org/licenses/gpl-3.0.en.html) + + +The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code. + +**MIScnn provides several core features:** +- 2D/3D medical image segmentation for binary and multi-class problems +- Data I/O, preprocessing and data augmentation for biomedical images +- Patch-wise and full image analysis +- State-of-the-art deep learning model and metric library +- Intuitive and fast model utilization (training, prediction) +- Multiple automatic evaluation techniques (e.g. cross-validation) +- Custom model, data I/O, pre-/postprocessing and metric support +- Based on Keras with Tensorflow as backend + +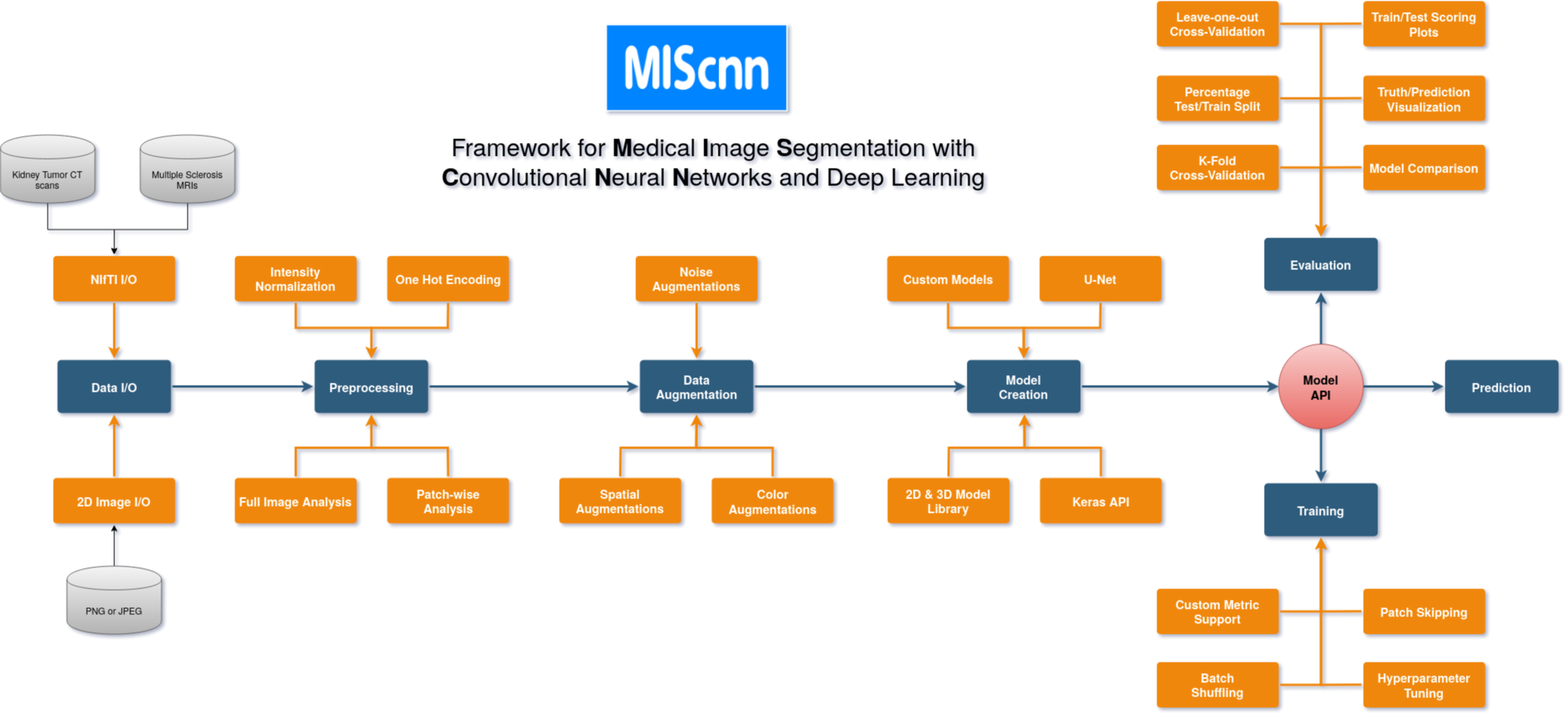 + +## Resources + +- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki) +- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials) +- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples) +- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1) +- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn) +- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/) + +## Author + +Dominik Müller +Email: dominik.mueller@informatik.uni-augsburg.de +IT-Infrastructure for Translational Medical Research +University Augsburg +Augsburg, Bavaria, Germany + +## How to cite / More information + +Dominik Müller and Frank Kramer. (2019) +MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning. +arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308) + +``` +Article{miscnn, + title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning}, + author={Dominik Müller and Frank Kramer}, + year={2019}, + eprint={1910.09308}, + archivePrefix={arXiv}, + primaryClass={eess.IV} +} +``` + +Thank you for citing our work. + +## License + +This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\ +See the LICENSE.md file for license rights and limitations. + + + + +%package -n python3-miscnn +Summary: Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning +Provides: python-miscnn +BuildRequires: python3-devel +BuildRequires: python3-setuptools +BuildRequires: python3-pip +%description -n python3-miscnn + + +[](https://www.python.org/) +[](https://travis-ci.org/github/frankkramer-lab/MIScnn) +[](https://codecov.io/gh/frankkramer-lab/miscnn) +[](https://pypi.org/project/miscnn/) +[](https://pypistats.org/packages/miscnn) +[](https://www.gnu.org/licenses/gpl-3.0.en.html) + + +The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code. + +**MIScnn provides several core features:** +- 2D/3D medical image segmentation for binary and multi-class problems +- Data I/O, preprocessing and data augmentation for biomedical images +- Patch-wise and full image analysis +- State-of-the-art deep learning model and metric library +- Intuitive and fast model utilization (training, prediction) +- Multiple automatic evaluation techniques (e.g. cross-validation) +- Custom model, data I/O, pre-/postprocessing and metric support +- Based on Keras with Tensorflow as backend + + + +## Resources + +- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki) +- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials) +- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples) +- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1) +- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn) +- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/) + +## Author + +Dominik Müller +Email: dominik.mueller@informatik.uni-augsburg.de +IT-Infrastructure for Translational Medical Research +University Augsburg +Augsburg, Bavaria, Germany + +## How to cite / More information + +Dominik Müller and Frank Kramer. (2019) +MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning. +arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308) + +``` +Article{miscnn, + title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning}, + author={Dominik Müller and Frank Kramer}, + year={2019}, + eprint={1910.09308}, + archivePrefix={arXiv}, + primaryClass={eess.IV} +} +``` + +Thank you for citing our work. + +## License + +This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\ +See the LICENSE.md file for license rights and limitations. + + + + +%package help +Summary: Development documents and examples for miscnn +Provides: python3-miscnn-doc +%description help + + +[](https://www.python.org/) +[](https://travis-ci.org/github/frankkramer-lab/MIScnn) +[](https://codecov.io/gh/frankkramer-lab/miscnn) +[](https://pypi.org/project/miscnn/) +[](https://pypistats.org/packages/miscnn) +[](https://www.gnu.org/licenses/gpl-3.0.en.html) + + +The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code. + +**MIScnn provides several core features:** +- 2D/3D medical image segmentation for binary and multi-class problems +- Data I/O, preprocessing and data augmentation for biomedical images +- Patch-wise and full image analysis +- State-of-the-art deep learning model and metric library +- Intuitive and fast model utilization (training, prediction) +- Multiple automatic evaluation techniques (e.g. cross-validation) +- Custom model, data I/O, pre-/postprocessing and metric support +- Based on Keras with Tensorflow as backend + + + +## Resources + +- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki) +- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials) +- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples) +- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1) +- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn) +- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/) + +## Author + +Dominik Müller +Email: dominik.mueller@informatik.uni-augsburg.de +IT-Infrastructure for Translational Medical Research +University Augsburg +Augsburg, Bavaria, Germany + +## How to cite / More information + +Dominik Müller and Frank Kramer. (2019) +MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning. +arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308) + +``` +Article{miscnn, + title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning}, + author={Dominik Müller and Frank Kramer}, + year={2019}, + eprint={1910.09308}, + archivePrefix={arXiv}, + primaryClass={eess.IV} +} +``` + +Thank you for citing our work. + +## License + +This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\ +See the LICENSE.md file for license rights and limitations. + + + + +%prep +%autosetup -n miscnn-1.4.0 + +%build +%py3_build + +%install +%py3_install +install -d -m755 %{buildroot}/%{_pkgdocdir} +if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi +if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi +if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi +if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi +pushd %{buildroot} +if [ -d usr/lib ]; then + find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/lib64 ]; then + find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/bin ]; then + find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst +fi +if [ -d usr/sbin ]; then + find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst +fi +touch doclist.lst +if [ -d usr/share/man ]; then + find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst +fi +popd +mv %{buildroot}/filelist.lst . +mv %{buildroot}/doclist.lst . + +%files -n python3-miscnn -f filelist.lst +%dir %{python3_sitelib}/* + +%files help -f doclist.lst +%{_docdir}/* + +%changelog +* Wed May 10 2023 Python_Bot <Python_Bot@openeuler.org> - 1.4.0-1 +- Package Spec generated @@ -0,0 +1 @@ +2fdc15f5407170795ef508c2c846de37 miscnn-1.4.0.tar.gz |
