1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
|
%global _empty_manifest_terminate_build 0
Name: python-miscnn
Version: 1.4.0
Release: 1
Summary: Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning
License: GPLv3
URL: https://github.com/frankkramer-lab/MIScnn
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/3e/c6/3368ea5168d440a809264a0815beb9b26516c137a1fb106c3596e0f5125e/miscnn-1.4.0.tar.gz
BuildArch: noarch
Requires: python3-tensorflow
Requires: python3-tensorflow-addons
Requires: python3-numpy
Requires: python3-pandas
Requires: python3-tqdm
Requires: python3-nibabel
Requires: python3-matplotlib
Requires: python3-pillow
Requires: python3-batchgenerators
Requires: python3-pydicom
Requires: python3-SimpleITK
Requires: python3-scikit-image
%description

[](https://www.python.org/)
[](https://travis-ci.org/github/frankkramer-lab/MIScnn)
[](https://codecov.io/gh/frankkramer-lab/miscnn)
[](https://pypi.org/project/miscnn/)
[](https://pypistats.org/packages/miscnn)
[](https://www.gnu.org/licenses/gpl-3.0.en.html)
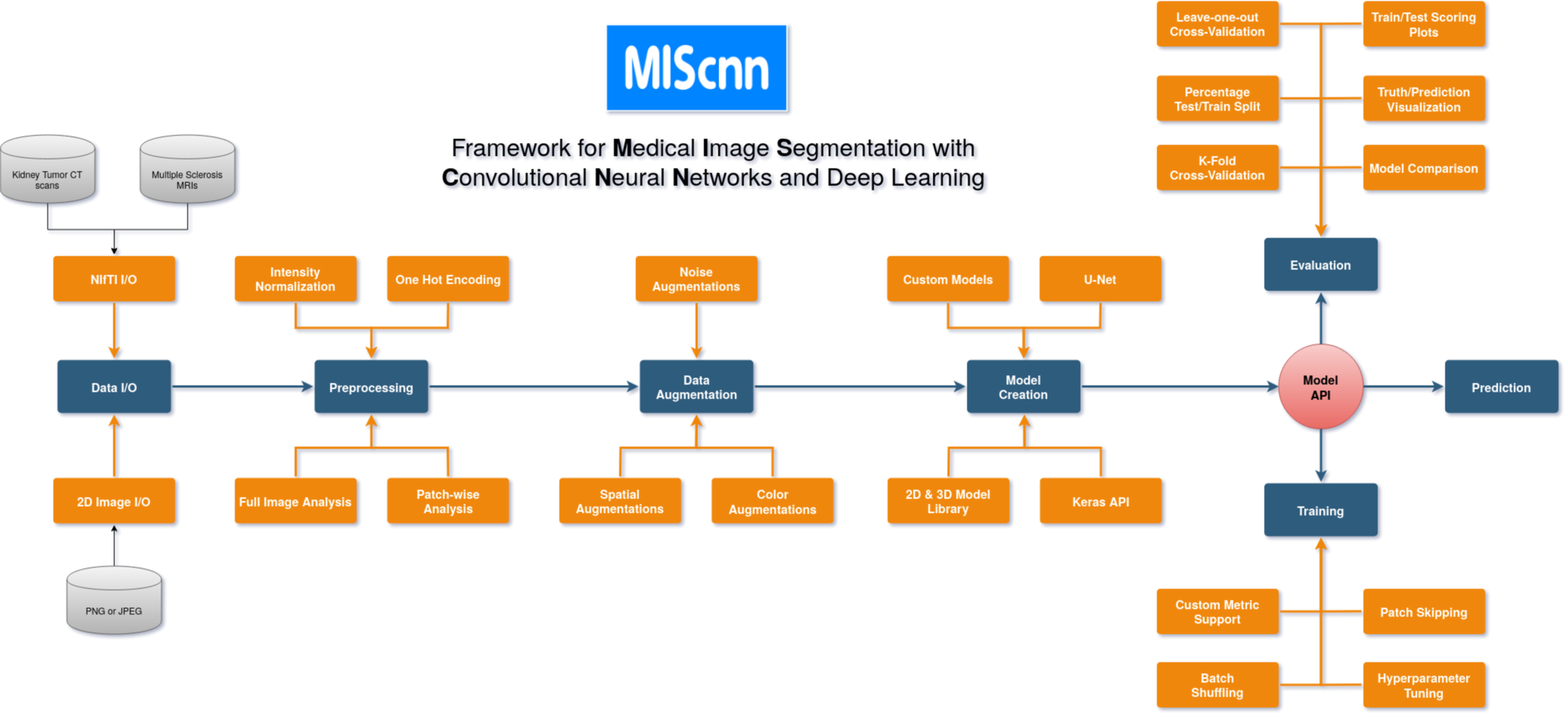
The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code.
**MIScnn provides several core features:**
- 2D/3D medical image segmentation for binary and multi-class problems
- Data I/O, preprocessing and data augmentation for biomedical images
- Patch-wise and full image analysis
- State-of-the-art deep learning model and metric library
- Intuitive and fast model utilization (training, prediction)
- Multiple automatic evaluation techniques (e.g. cross-validation)
- Custom model, data I/O, pre-/postprocessing and metric support
- Based on Keras with Tensorflow as backend
## Resources
- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki)
- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials)
- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples)
- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1)
- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn)
- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/)
## Author
Dominik Müller
Email: dominik.mueller@informatik.uni-augsburg.de
IT-Infrastructure for Translational Medical Research
University Augsburg
Augsburg, Bavaria, Germany
## How to cite / More information
Dominik Müller and Frank Kramer. (2019)
MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning.
arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308)
```
Article{miscnn,
title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning},
author={Dominik Müller and Frank Kramer},
year={2019},
eprint={1910.09308},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
```
Thank you for citing our work.
## License
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\
See the LICENSE.md file for license rights and limitations.
%package -n python3-miscnn
Summary: Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning
Provides: python-miscnn
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-miscnn

[](https://www.python.org/)
[](https://travis-ci.org/github/frankkramer-lab/MIScnn)
[](https://codecov.io/gh/frankkramer-lab/miscnn)
[](https://pypi.org/project/miscnn/)
[](https://pypistats.org/packages/miscnn)
[](https://www.gnu.org/licenses/gpl-3.0.en.html)
The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code.
**MIScnn provides several core features:**
- 2D/3D medical image segmentation for binary and multi-class problems
- Data I/O, preprocessing and data augmentation for biomedical images
- Patch-wise and full image analysis
- State-of-the-art deep learning model and metric library
- Intuitive and fast model utilization (training, prediction)
- Multiple automatic evaluation techniques (e.g. cross-validation)
- Custom model, data I/O, pre-/postprocessing and metric support
- Based on Keras with Tensorflow as backend

## Resources
- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki)
- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials)
- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples)
- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1)
- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn)
- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/)
## Author
Dominik Müller
Email: dominik.mueller@informatik.uni-augsburg.de
IT-Infrastructure for Translational Medical Research
University Augsburg
Augsburg, Bavaria, Germany
## How to cite / More information
Dominik Müller and Frank Kramer. (2019)
MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning.
arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308)
```
Article{miscnn,
title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning},
author={Dominik Müller and Frank Kramer},
year={2019},
eprint={1910.09308},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
```
Thank you for citing our work.
## License
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\
See the LICENSE.md file for license rights and limitations.
%package help
Summary: Development documents and examples for miscnn
Provides: python3-miscnn-doc
%description help

[](https://www.python.org/)
[](https://travis-ci.org/github/frankkramer-lab/MIScnn)
[](https://codecov.io/gh/frankkramer-lab/miscnn)
[](https://pypi.org/project/miscnn/)
[](https://pypistats.org/packages/miscnn)
[](https://www.gnu.org/licenses/gpl-3.0.en.html)
The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code.
**MIScnn provides several core features:**
- 2D/3D medical image segmentation for binary and multi-class problems
- Data I/O, preprocessing and data augmentation for biomedical images
- Patch-wise and full image analysis
- State-of-the-art deep learning model and metric library
- Intuitive and fast model utilization (training, prediction)
- Multiple automatic evaluation techniques (e.g. cross-validation)
- Custom model, data I/O, pre-/postprocessing and metric support
- Based on Keras with Tensorflow as backend

## Resources
- MIScnn Documentation: [GitHub wiki - Home](https://github.com/frankkramer-lab/MIScnn/wiki)
- MIScnn Tutorials: [Overview of Tutorials](https://github.com/frankkramer-lab/MIScnn/wiki/Tutorials)
- MIScnn Examples: [Overview of Use Cases and Examples](https://github.com/frankkramer-lab/MIScnn/wiki/Examples)
- MIScnn Development Tracker: [GitHub project - MIScnn Development](https://github.com/frankkramer-lab/MIScnn/projects/1)
- MIScnn on GitHub: [GitHub - frankkramer-lab/MIScnn](https://github.com/frankkramer-lab/MIScnn)
- MIScnn on PyPI: [PyPI - miscnn](https://pypi.org/project/miscnn/)
## Author
Dominik Müller
Email: dominik.mueller@informatik.uni-augsburg.de
IT-Infrastructure for Translational Medical Research
University Augsburg
Augsburg, Bavaria, Germany
## How to cite / More information
Dominik Müller and Frank Kramer. (2019)
MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning.
arXiv e-print: [https://arxiv.org/abs/1910.09308](https://arxiv.org/abs/1910.09308)
```
Article{miscnn,
title={MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning},
author={Dominik Müller and Frank Kramer},
year={2019},
eprint={1910.09308},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
```
Thank you for citing our work.
## License
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.\
See the LICENSE.md file for license rights and limitations.
%prep
%autosetup -n miscnn-1.4.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-miscnn -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed May 10 2023 Python_Bot <Python_Bot@openeuler.org> - 1.4.0-1
- Package Spec generated
|
