%global _empty_manifest_terminate_build 0
Name: python-pmdarima
Version: 2.0.3
Release: 1
Summary: Python's forecast::auto.arima equivalent
License: MIT
URL: http://alkaline-ml.com/pmdarima
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/61/05/0690150bb6784db4d30fcde6062e9410597f767f024a0d1a5fd850804ec9/pmdarima-2.0.3.tar.gz
Requires: python3-joblib
Requires: python3-Cython
Requires: python3-numpy
Requires: python3-pandas
Requires: python3-scikit-learn
Requires: python3-scipy
Requires: python3-statsmodels
Requires: python3-urllib3
Requires: python3-setuptools
%description
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
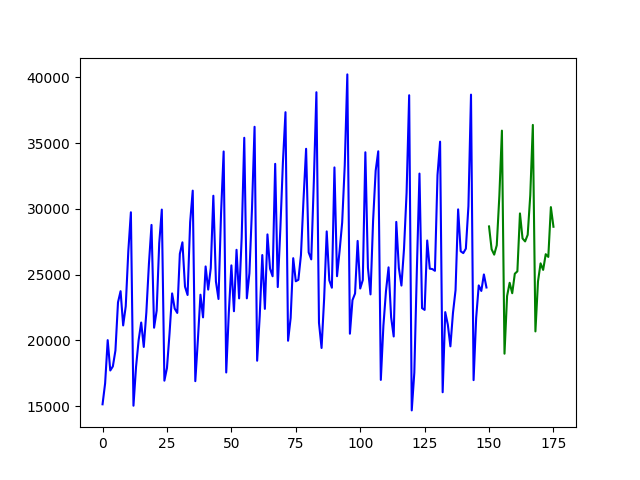
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package -n python3-pmdarima
Summary: Python's forecast::auto.arima equivalent
Provides: python-pmdarima
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
BuildRequires: python3-cffi
BuildRequires: gcc
BuildRequires: gdb
%description -n python3-pmdarima
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package -n python3-pmdarima
Summary: Python's forecast::auto.arima equivalent
Provides: python-pmdarima
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
BuildRequires: python3-cffi
BuildRequires: gcc
BuildRequires: gdb
%description -n python3-pmdarima
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package help
Summary: Development documents and examples for pmdarima
Provides: python3-pmdarima-doc
%description help
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package help
Summary: Development documents and examples for pmdarima
Provides: python3-pmdarima-doc
%description help
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%prep
%autosetup -n pmdarima-2.0.3
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pmdarima -f filelist.lst
%dir %{python3_sitearch}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Mon Apr 10 2023 Python_Bot - 2.0.3-1
- Package Spec generated
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%prep
%autosetup -n pmdarima-2.0.3
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pmdarima -f filelist.lst
%dir %{python3_sitearch}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Mon Apr 10 2023 Python_Bot - 2.0.3-1
- Package Spec generated
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package -n python3-pmdarima
Summary: Python's forecast::auto.arima equivalent
Provides: python-pmdarima
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
BuildRequires: python3-cffi
BuildRequires: gcc
BuildRequires: gdb
%description -n python3-pmdarima
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package -n python3-pmdarima
Summary: Python's forecast::auto.arima equivalent
Provides: python-pmdarima
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
BuildRequires: python3-cffi
BuildRequires: gcc
BuildRequires: gdb
%description -n python3-pmdarima
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package help
Summary: Development documents and examples for pmdarima
Provides: python3-pmdarima-doc
%description help
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%package help
Summary: Development documents and examples for pmdarima
Provides: python3-pmdarima-doc
%description help
# pmdarima
[](https://badge.fury.io/py/pmdarima)
[](https://circleci.com/gh/alkaline-ml/pmdarima)
[](https://github.com/alkaline-ml/pmdarima/actions?query=workflow%3A%22Mac+and+Windows+Builds%22+branch%3Amaster)
[](https://codecov.io/gh/alkaline-ml/pmdarima)



Pmdarima (originally `pyramid-arima`, for the anagram of 'py' + 'arima') is a statistical
library designed to fill the void in Python's time series analysis capabilities. This includes:
* The equivalent of R's [`auto.arima`](https://www.rdocumentation.org/packages/forecast/versions/7.3/topics/auto.arima) functionality
* A collection of statistical tests of stationarity and seasonality
* Time series utilities, such as differencing and inverse differencing
* Numerous endogenous and exogenous transformers and featurizers, including Box-Cox and Fourier transformations
* Seasonal time series decompositions
* Cross-validation utilities
* A rich collection of built-in time series datasets for prototyping and examples
* Scikit-learn-esque pipelines to consolidate your estimators and promote productionization
Pmdarima wraps [statsmodels](https://github.com/statsmodels/statsmodels/blob/master/statsmodels)
under the hood, but is designed with an interface that's familiar to users coming
from a scikit-learn background.
## Installation
### pip
Pmdarima has binary and source distributions for Windows, Mac and Linux (`manylinux`) on pypi
under the package name `pmdarima` and can be downloaded via `pip`:
```bash
pip install pmdarima
```
### conda
Pmdarima also has Mac and Linux builds available via `conda` and can be installed like so:
```bash
conda config --add channels conda-forge
conda config --set channel_priority strict
conda install pmdarima
```
**Note:** We do not maintain our own Conda binaries, they are maintained at https://github.com/conda-forge/pmdarima-feedstock.
See that repo for further documentation on working with Pmdarima on Conda.
## Quickstart Examples
Fitting a simple auto-ARIMA on the [`wineind`](https://alkaline-ml.com/pmdarima/modules/generated/pmdarima.datasets.load_wineind.html#pmdarima.datasets.load_wineind) dataset:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
import numpy as np
import matplotlib.pyplot as plt
# Load/split your data
y = pm.datasets.load_wineind()
train, test = train_test_split(y, train_size=150)
# Fit your model
model = pm.auto_arima(train, seasonal=True, m=12)
# make your forecasts
forecasts = model.predict(test.shape[0]) # predict N steps into the future
# Visualize the forecasts (blue=train, green=forecasts)
x = np.arange(y.shape[0])
plt.plot(x[:150], train, c='blue')
plt.plot(x[150:], forecasts, c='green')
plt.show()
```
 Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%prep
%autosetup -n pmdarima-2.0.3
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pmdarima -f filelist.lst
%dir %{python3_sitearch}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Mon Apr 10 2023 Python_Bot
Fitting a more complex pipeline on the [`sunspots`](https://www.rdocumentation.org/packages/datasets/versions/3.6.1/topics/sunspots) dataset,
serializing it, and then loading it from disk to make predictions:
```python
import pmdarima as pm
from pmdarima.model_selection import train_test_split
from pmdarima.pipeline import Pipeline
from pmdarima.preprocessing import BoxCoxEndogTransformer
import pickle
# Load/split your data
y = pm.datasets.load_sunspots()
train, test = train_test_split(y, train_size=2700)
# Define and fit your pipeline
pipeline = Pipeline([
('boxcox', BoxCoxEndogTransformer(lmbda2=1e-6)), # lmbda2 avoids negative values
('arima', pm.AutoARIMA(seasonal=True, m=12,
suppress_warnings=True,
trace=True))
])
pipeline.fit(train)
# Serialize your model just like you would in scikit:
with open('model.pkl', 'wb') as pkl:
pickle.dump(pipeline, pkl)
# Load it and make predictions seamlessly:
with open('model.pkl', 'rb') as pkl:
mod = pickle.load(pkl)
print(mod.predict(15))
# [25.20580375 25.05573898 24.4263037 23.56766793 22.67463049 21.82231043
# 21.04061069 20.33693017 19.70906027 19.1509862 18.6555793 18.21577243
# 17.8250318 17.47750614 17.16803394]
```
### Availability
`pmdarima` is available on PyPi in pre-built Wheel files for Python 3.7+ for the following platforms:
* Mac (64-bit)
* Linux (64-bit manylinux)
* Windows (64-bit)
* 32-bit wheels are available for pmdarima versions below 2.0.0 and Python versions below 3.10
If a wheel doesn't exist for your platform, you can still `pip install` and it
will build from the source distribution tarball, however you'll need `cython>=0.29`
and `gcc` (Mac/Linux) or `MinGW` (Windows) in order to build the package from source.
Note that legacy versions (<1.0.0) are available under the name
"`pyramid-arima`" and can be pip installed via:
```bash
# Legacy warning:
$ pip install pyramid-arima
# python -c 'import pyramid;'
```
However, this is not recommended.
## Documentation
All of your questions and more (including examples and guides) can be answered by
the [`pmdarima` documentation](https://www.alkaline-ml.com/pmdarima). If not, always
feel free to file an issue.
%prep
%autosetup -n pmdarima-2.0.3
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pmdarima -f filelist.lst
%dir %{python3_sitearch}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Mon Apr 10 2023 Python_Bot