%global _empty_manifest_terminate_build 0
Name: python-pyxtal
Version: 0.5.5
Release: 1
Summary: Python code for generation of crystal structures based on symmetry constraints.
License: MIT
URL: https://github.com/qzhu2017/PyXtal
Source0: https://mirrors.aliyun.com/pypi/web/packages/b9/0f/c22a5925963b88bbb2ea7f7969e5446ae7482442ad967e99adad7d45fa26/pyxtal-0.5.5.tar.gz
BuildArch: noarch
Requires: python3-spglib
Requires: python3-pymatgen
Requires: python3-pandas
Requires: python3-networkx
Requires: python3-py3Dmol
Requires: python3-ase
Requires: python3-numba
Requires: python3-scipy
Requires: python3-importlib-metadata
Requires: python3-pyshtools
%description
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
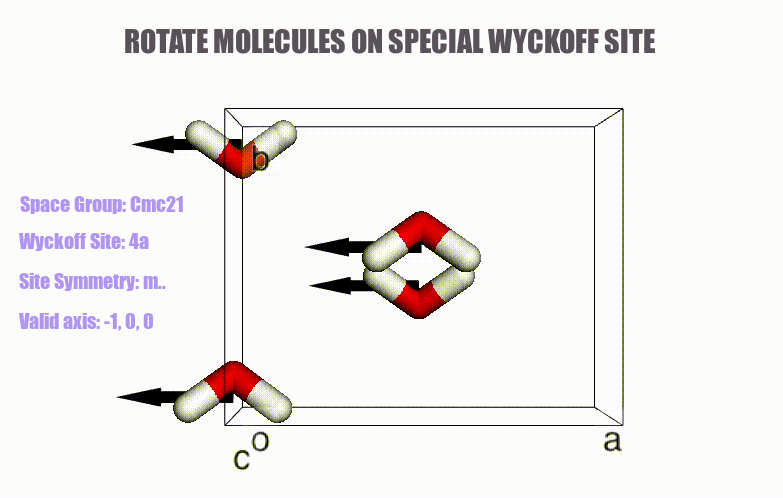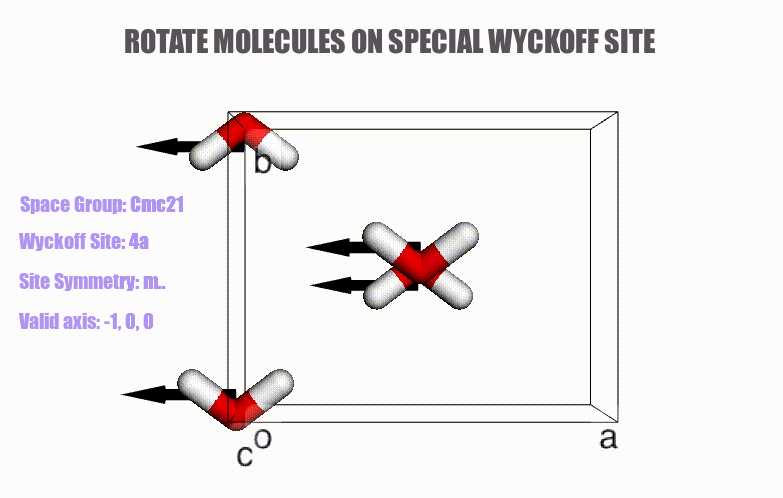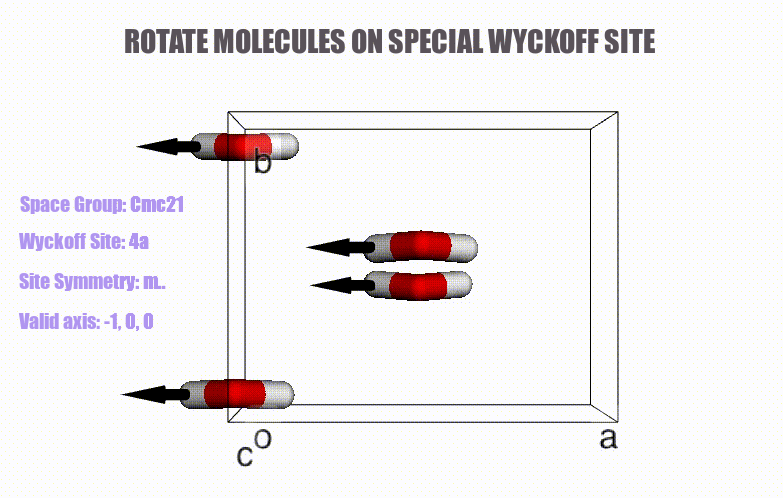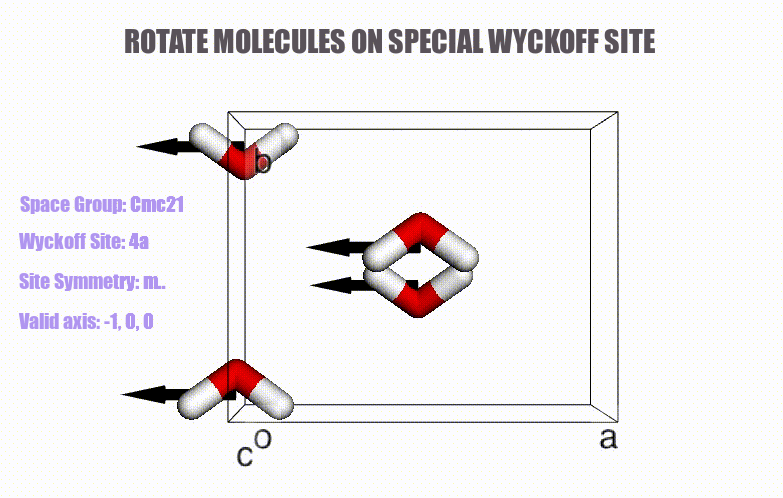 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package -n python3-pyxtal
Summary: Python code for generation of crystal structures based on symmetry constraints.
Provides: python-pyxtal
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-pyxtal
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package -n python3-pyxtal
Summary: Python code for generation of crystal structures based on symmetry constraints.
Provides: python-pyxtal
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-pyxtal
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package help
Summary: Development documents and examples for pyxtal
Provides: python3-pyxtal-doc
%description help
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package help
Summary: Development documents and examples for pyxtal
Provides: python3-pyxtal-doc
%description help
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%prep
%autosetup -n pyxtal-0.5.5
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pyxtal -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Thu Jun 08 2023 Python_Bot - 0.5.5-1
- Package Spec generated
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%prep
%autosetup -n pyxtal-0.5.5
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pyxtal -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Thu Jun 08 2023 Python_Bot - 0.5.5-1
- Package Spec generated
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package -n python3-pyxtal
Summary: Python code for generation of crystal structures based on symmetry constraints.
Provides: python-pyxtal
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-pyxtal
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package -n python3-pyxtal
Summary: Python code for generation of crystal structures based on symmetry constraints.
Provides: python-pyxtal
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-pyxtal
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package help
Summary: Development documents and examples for pyxtal
Provides: python3-pyxtal-doc
%description help
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%package help
Summary: Development documents and examples for pyxtal
Provides: python3-pyxtal-doc
%description help
 [](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
[](https://pyxtal.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/qzhu2017/PyXtal/actions)
[](https://pypi.org/project/pyxtal/)
[](https://pypi.org/project/pyxtal/)
[](https://pepy.tech/project/pyxtal)
[](https://zenodo.org/badge/latestdoi/128165891)
[](https://gitter.im/PyXtal/community?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge)
 ## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%prep
%autosetup -n pyxtal-0.5.5
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pyxtal -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Thu Jun 08 2023 Python_Bot
## Table of content
- [Introduction](#introduction)
- [Quick Start](#quick-start)
- [Current Features](#current-features)
- [Installation](#installation)
- [Citation](#citation)
- [How to contribute?](#how-to-contribute)
## Introduction
PyXtal is an open source Python package which was initiated by [Qiang Zhu](http://qzhu2017.github.io) and Scott Fredericks at department of Physics and Astronomy, University of Nevada Las Vegas. The goal of PyXtal is to develop a fundamental library to allow one to design the material structure with a certain symmetry constraint. These structures can exported to various structural formats for further study. See the [documentation](https://pyxtal.readthedocs.io/en/latest/) for more information.
To contribute to this project, please check [How to contribute?](#how-to-contribute).
## Quick Start
- [Atomic crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/01_atomic_crystals.ipynb)
- [Molecular crystal](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/02_molecular_crystals.ipynb)
- [XRD](https://nbviewer.jupyter.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/03_pxrd.ipynb)
- [Molecular Packing](https://nbviewer.org/github/qzhu2017/PyXtal/blob/master/examples/tutorials_notebook/05-crystal-packing.ipynb)
## Current Features
- Generation of atomic structures for a given symmetry and stoichiometry (0-3D)
- Generation of molecular crystals (1-3D) with the support of special Wyckoff positions.
- Structure manipulation via subgroup/supergroup symmetry relation
- Geometry optimization from built-in and external optimization methods.
- Internal support of ``cif`` file and many other formats via ``pymatgen`` or ``ASE``.
- Easy access to symmetry information (e.g., Wyckoff, site symmetry and international symbols).
- X-ray diffraction analysis and [its online application](https://vxrd.physics.unlv.edu)
## Installation
To install the code, one just needs to do
```sh
pip install pyxtal
```
or
```sh
pip install --upgrade git+https://github.com/qzhu2017/PyXtal.git@master
```
## Citation
Fredericks S, Parrish K, Sayre D, Zhu Q\*(2020)
[PyXtal: a Python Library for Crystal Structure Generation and Symmetry Analysis](https://www.sciencedirect.com/science/article/pii/S0010465520304057).
\[[arXiv link](https://arxiv.org/pdf/1911.11123.pdf)\]
```bib
@article{pyxtal,
title = "PyXtal: A Python library for crystal structure generation and symmetry analysis",
journal = "Computer Physics Communications",
volume = "261",
pages = "107810",
year = "2021",
issn = "0010-4655",
doi = "https://doi.org/10.1016/j.cpc.2020.107810",
url = "http://www.sciencedirect.com/science/article/pii/S0010465520304057",
author = "Scott Fredericks and Kevin Parrish and Dean Sayre and Qiang Zhu",
}
```
## How to contribute?
This is an open-source project. Its growth depends on the community. To contribute to PyXtal, you don't necessarily have to write the code. Any contributions from the following list will be helpful.
- [](https://github.com/qzhu2017/pyxtal/stargazers)
the PyXtal project and recommend it to your colleagues/friends
- Open an [](https://GitHub.com/qzhu2017/pyxtal/issues/) to report the bug or address your wishlist or improve our documentation
- [](https://github.com/qzhu2017/PyXtal/network/members) the repository and send us the pull request
%prep
%autosetup -n pyxtal-0.5.5
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-pyxtal -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Thu Jun 08 2023 Python_Bot