%global _empty_manifest_terminate_build 0
Name: python-wfdb
Version: 4.1.0
Release: 1
Summary: The WFDB Python package: tools for reading, writing, and processing physiologic signals and annotations.
License: MIT
URL: https://github.com/MIT-LCP/wfdb-python/
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/e8/e2/e78d4322eaeabf3309c966e69e7cd64c1107638d86ba47d711099f240383/wfdb-4.1.0.tar.gz
BuildArch: noarch
Requires: python3-SoundFile
Requires: python3-Sphinx
Requires: python3-black
Requires: python3-matplotlib
Requires: python3-numpy
Requires: python3-pandas
Requires: python3-pylint
Requires: python3-pytest
Requires: python3-pytest-xdist
Requires: python3-requests
Requires: python3-scipy
%description
# The WFDB Python Package
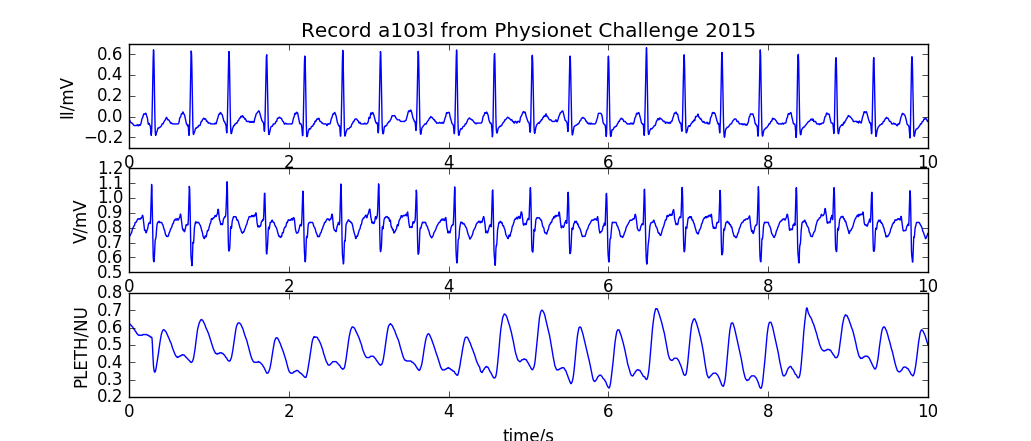
[](https://github.com/MIT-LCP/wfdb-python/actions?query=workflow%3Arun-tests+event%3Apush+branch%3Amain)
[](https://pypi.org/project/wfdb/)
[](https://doi.org/10.13026/egpf-2788)
[](https://pypi.org/project/wfdb)
## Introduction
A Python-native package for reading, writing, processing, and plotting physiologic signal and annotation data. The core I/O functionality is based on the Waveform Database (WFDB) [specifications](https://github.com/wfdb/wfdb-spec/).
This package is heavily inspired by the original [WFDB Software Package](https://www.physionet.org/content/wfdb/), and initially aimed to replicate many of its command-line APIs. However, the projects are independent, and there is no promise of consistency between the two, beyond each package adhering to the core specifications.
## Documentation and Usage
See the [documentation site](http://wfdb.readthedocs.io) for the public APIs.
See the [demo.ipynb](https://github.com/MIT-LCP/wfdb-python/blob/main/demo.ipynb) notebook file for example use cases.
## Installation
The distribution is hosted on PyPI at: . The package can be directly installed from PyPI using either pip or poetry:
```sh
pip install wfdb
poetry add wfdb
```
On Linux systems, accessing _compressed_ WFDB signal files requires installing `libsndfile`, by running `sudo apt-get install libsndfile1` or `sudo yum install libsndfile`. Support for Apple M1 systems is a work in progess (see and ).
The development version is hosted at: . This repository also contains demo scripts and example data. To install the development version, clone or download the repository, navigate to the base directory, and run:
```sh
# Without dev dependencies
pip install .
poetry install
# With dev dependencies
pip install ".[dev]"
poetry install -E dev
# Install the dependencies only
poetry install -E dev --no-root
```
**See the [note](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md#package-and-dependency-management) about dev dependencies.**
## Developing
Please see the [DEVELOPING.md](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md) document for contribution/development instructions.
## Citing
When using this resource, please cite the software [publication](https://physionet.org/content/wfdb-python/) on PhysioNet.
%package -n python3-wfdb
Summary: The WFDB Python package: tools for reading, writing, and processing physiologic signals and annotations.
Provides: python-wfdb
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-wfdb
# The WFDB Python Package

[](https://github.com/MIT-LCP/wfdb-python/actions?query=workflow%3Arun-tests+event%3Apush+branch%3Amain)
[](https://pypi.org/project/wfdb/)
[](https://doi.org/10.13026/egpf-2788)
[](https://pypi.org/project/wfdb)
## Introduction
A Python-native package for reading, writing, processing, and plotting physiologic signal and annotation data. The core I/O functionality is based on the Waveform Database (WFDB) [specifications](https://github.com/wfdb/wfdb-spec/).
This package is heavily inspired by the original [WFDB Software Package](https://www.physionet.org/content/wfdb/), and initially aimed to replicate many of its command-line APIs. However, the projects are independent, and there is no promise of consistency between the two, beyond each package adhering to the core specifications.
## Documentation and Usage
See the [documentation site](http://wfdb.readthedocs.io) for the public APIs.
See the [demo.ipynb](https://github.com/MIT-LCP/wfdb-python/blob/main/demo.ipynb) notebook file for example use cases.
## Installation
The distribution is hosted on PyPI at: . The package can be directly installed from PyPI using either pip or poetry:
```sh
pip install wfdb
poetry add wfdb
```
On Linux systems, accessing _compressed_ WFDB signal files requires installing `libsndfile`, by running `sudo apt-get install libsndfile1` or `sudo yum install libsndfile`. Support for Apple M1 systems is a work in progess (see and ).
The development version is hosted at: . This repository also contains demo scripts and example data. To install the development version, clone or download the repository, navigate to the base directory, and run:
```sh
# Without dev dependencies
pip install .
poetry install
# With dev dependencies
pip install ".[dev]"
poetry install -E dev
# Install the dependencies only
poetry install -E dev --no-root
```
**See the [note](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md#package-and-dependency-management) about dev dependencies.**
## Developing
Please see the [DEVELOPING.md](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md) document for contribution/development instructions.
## Citing
When using this resource, please cite the software [publication](https://physionet.org/content/wfdb-python/) on PhysioNet.
%package help
Summary: Development documents and examples for wfdb
Provides: python3-wfdb-doc
%description help
# The WFDB Python Package

[](https://github.com/MIT-LCP/wfdb-python/actions?query=workflow%3Arun-tests+event%3Apush+branch%3Amain)
[](https://pypi.org/project/wfdb/)
[](https://doi.org/10.13026/egpf-2788)
[](https://pypi.org/project/wfdb)
## Introduction
A Python-native package for reading, writing, processing, and plotting physiologic signal and annotation data. The core I/O functionality is based on the Waveform Database (WFDB) [specifications](https://github.com/wfdb/wfdb-spec/).
This package is heavily inspired by the original [WFDB Software Package](https://www.physionet.org/content/wfdb/), and initially aimed to replicate many of its command-line APIs. However, the projects are independent, and there is no promise of consistency between the two, beyond each package adhering to the core specifications.
## Documentation and Usage
See the [documentation site](http://wfdb.readthedocs.io) for the public APIs.
See the [demo.ipynb](https://github.com/MIT-LCP/wfdb-python/blob/main/demo.ipynb) notebook file for example use cases.
## Installation
The distribution is hosted on PyPI at: . The package can be directly installed from PyPI using either pip or poetry:
```sh
pip install wfdb
poetry add wfdb
```
On Linux systems, accessing _compressed_ WFDB signal files requires installing `libsndfile`, by running `sudo apt-get install libsndfile1` or `sudo yum install libsndfile`. Support for Apple M1 systems is a work in progess (see and ).
The development version is hosted at: . This repository also contains demo scripts and example data. To install the development version, clone or download the repository, navigate to the base directory, and run:
```sh
# Without dev dependencies
pip install .
poetry install
# With dev dependencies
pip install ".[dev]"
poetry install -E dev
# Install the dependencies only
poetry install -E dev --no-root
```
**See the [note](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md#package-and-dependency-management) about dev dependencies.**
## Developing
Please see the [DEVELOPING.md](https://github.com/MIT-LCP/wfdb-python/blob/main/DEVELOPING.md) document for contribution/development instructions.
## Citing
When using this resource, please cite the software [publication](https://physionet.org/content/wfdb-python/) on PhysioNet.
%prep
%autosetup -n wfdb-4.1.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-wfdb -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed Apr 12 2023 Python_Bot - 4.1.0-1
- Package Spec generated