%global _empty_manifest_terminate_build 0
Name: python-string-kernels
Version: 1.1.2
Release: 1
Summary: Polynomial String Kernel and linear time String Kernel. Supports multithreading and is compatible with Scikit-Learn SVMs.
License: Academic Free License (AFL)
URL: https://github.com/weekend37/string-kernels
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/ff/e7/04765217cdeced887f495c6833a93227d89f8342b1254f8979d294cb8874/string-kernels-1.1.2.tar.gz
BuildArch: noarch
%description
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
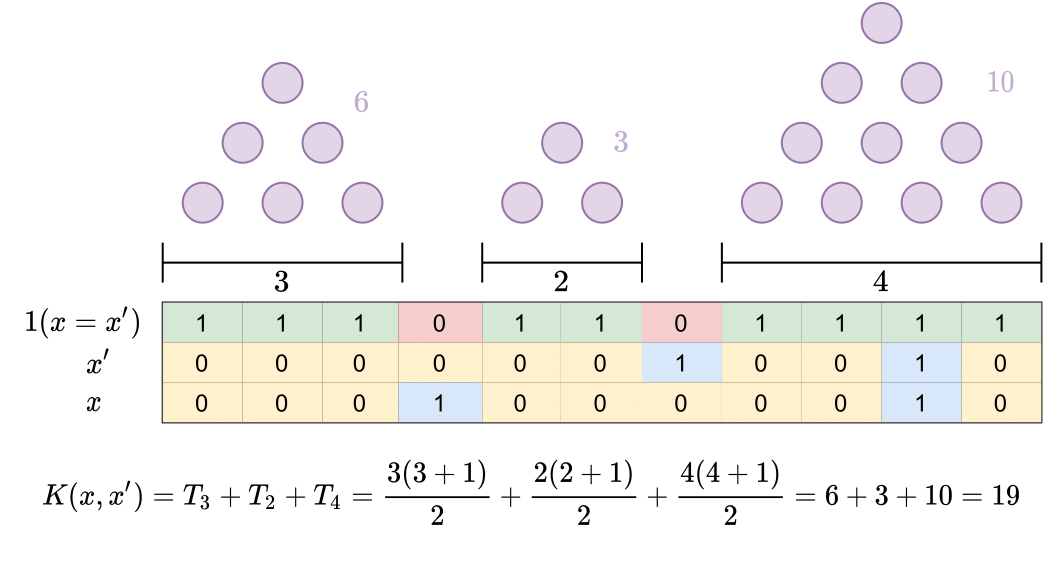 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package -n python3-string-kernels
Summary: Polynomial String Kernel and linear time String Kernel. Supports multithreading and is compatible with Scikit-Learn SVMs.
Provides: python-string-kernels
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-string-kernels
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package -n python3-string-kernels
Summary: Polynomial String Kernel and linear time String Kernel. Supports multithreading and is compatible with Scikit-Learn SVMs.
Provides: python-string-kernels
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-string-kernels
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
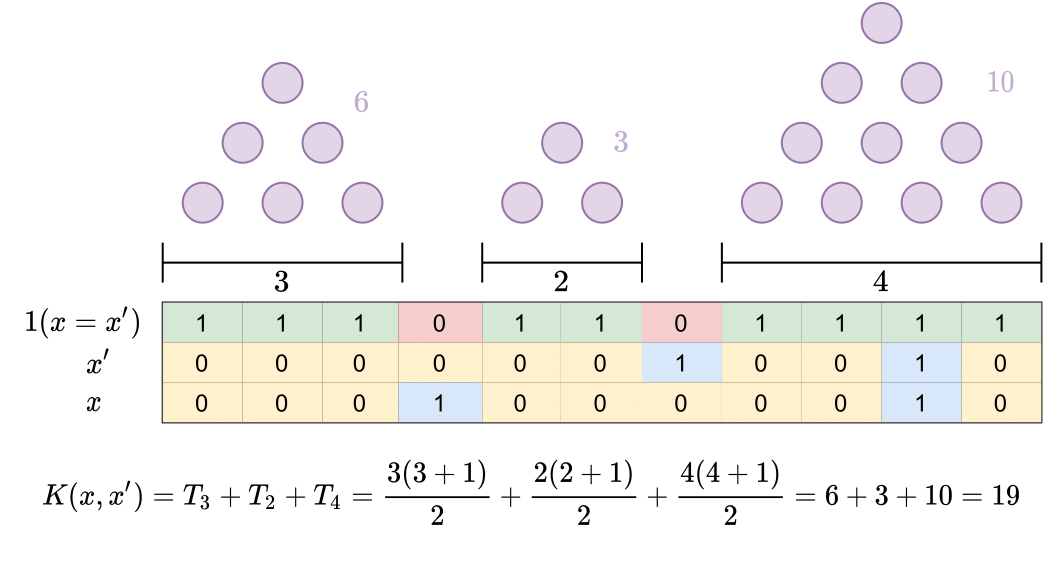 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package help
Summary: Development documents and examples for string-kernels
Provides: python3-string-kernels-doc
%description help
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package help
Summary: Development documents and examples for string-kernels
Provides: python3-string-kernels-doc
%description help
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
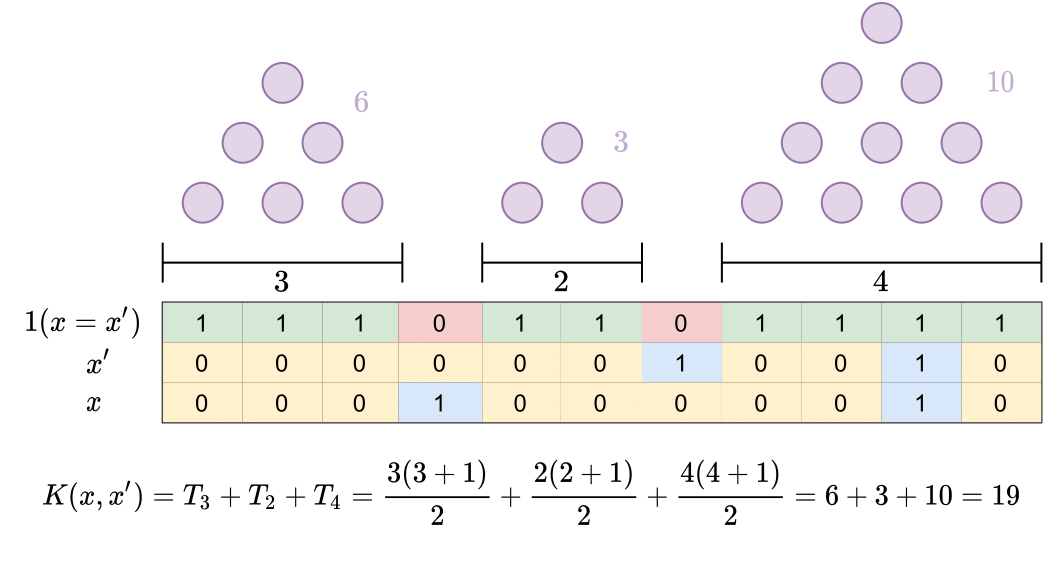 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%prep
%autosetup -n string-kernels-1.1.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-string-kernels -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed May 31 2023 Python_Bot - 1.1.2-1
- Package Spec generated
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%prep
%autosetup -n string-kernels-1.1.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-string-kernels -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed May 31 2023 Python_Bot - 1.1.2-1
- Package Spec generated
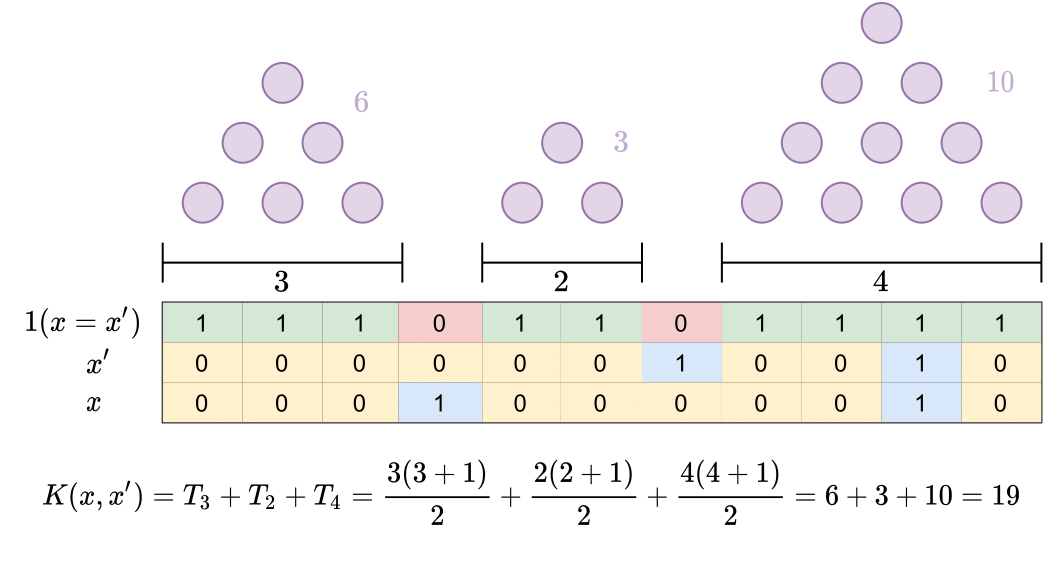 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package -n python3-string-kernels
Summary: Polynomial String Kernel and linear time String Kernel. Supports multithreading and is compatible with Scikit-Learn SVMs.
Provides: python-string-kernels
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-string-kernels
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package -n python3-string-kernels
Summary: Polynomial String Kernel and linear time String Kernel. Supports multithreading and is compatible with Scikit-Learn SVMs.
Provides: python-string-kernels
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-string-kernels
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1). 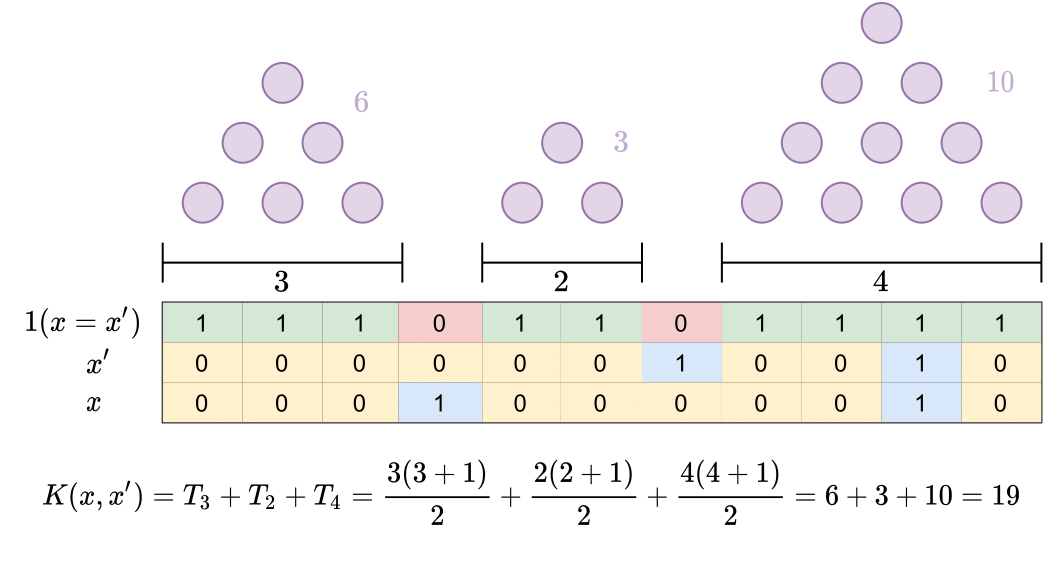 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package help
Summary: Development documents and examples for string-kernels
Provides: python3-string-kernels-doc
%description help
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1).
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%package help
Summary: Development documents and examples for string-kernels
Provides: python3-string-kernels-doc
%description help
# String Kernels
This package contains an implementation of the **Polynomial String Kernel** and a linear time **String Kernel** algorithm as described in our paper, [High Resolution Ancestry Deconvolution for Next Generation Genomic Data](https://www.biorxiv.org/content/10.1101/2021.09.19.460980v1). 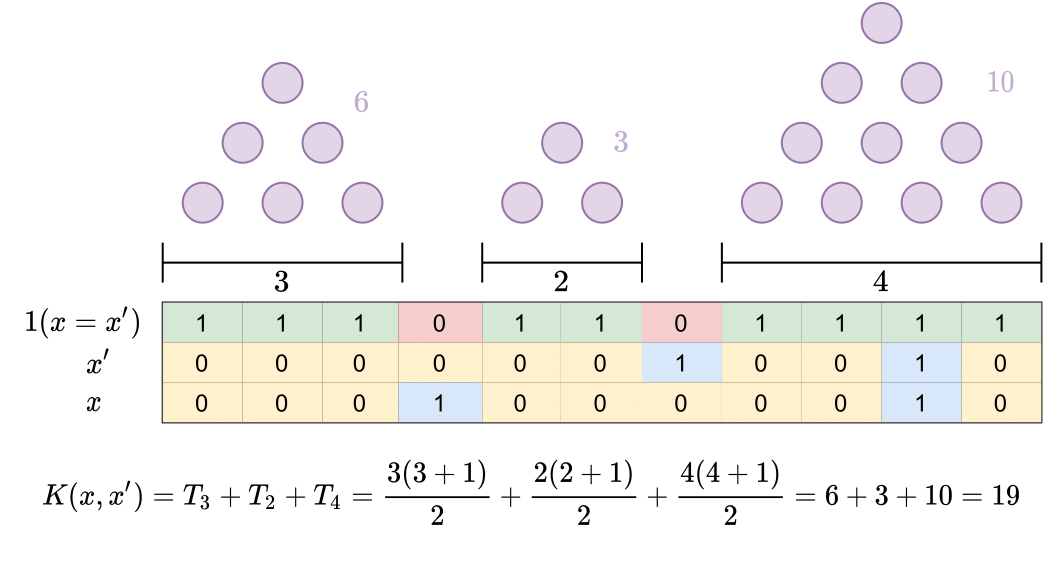 It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%prep
%autosetup -n string-kernels-1.1.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-string-kernels -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed May 31 2023 Python_Bot
It offers
- Linear time computation of two effective string kernels.
- Compatibility with Scikit-Learn's Support Vector Machines (easy plug-in).
- Multithreading.
## Usage
To install the package, execute from the command line
```
pip install string-kernels
```
And then you're all set!
Assuming you have [Scikit-Learn](https://scikit-learn.org/) already installed, you can use Lodhi's string kernel via
```python
from sklearn import svm
from stringkernels.kernels import string_kernel
model = svm.SVC(kernel=string_kernel())
```
and the polynomial string kernel,
```python
from sklearn import svm
from stringkernels.kernels import polynomial_string_kernel
model = svm.SVC(kernel=polynomial_string_kernel())
```
For morer information read the [docs](https://github.com/weekend37/string-kernels/blob/master/doc/docs.md) or take a look at the notebook [example.ipynb](https://github.com/weekend37/string-kernels/blob/master/example.ipynb) for further demonstration of usage.
If you end up using this in your research we kindly ask you to cite us! :)
%prep
%autosetup -n string-kernels-1.1.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-string-kernels -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Wed May 31 2023 Python_Bot