%global _empty_manifest_terminate_build 0
Name: python-cmaes
Version: 0.9.1
Release: 1
Summary: Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) implementation for Python 3.
License: MIT License Copyright (c) 2020-2021 CyberAgent, Inc. Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
URL: https://github.com/CyberAgentAILab/cmaes
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/d1/f9/35a3c4d887014b8464b4bccb364512fb0e87efb78bace58ec59f1efead63/cmaes-0.9.1.tar.gz
BuildArch: noarch
Requires: python3-numpy
Requires: python3-scipy
%description
# CMA-ES
[](./LICENSE) [](https://pypistats.org/packages/cmaes)
Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) [1] implementation.

## News
* **2022/05/13** The paper, ["CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization"](https://arxiv.org/abs/2205.13482) written by Hamano, Saito, [@nomuramasahir0](https://github.com/nomuramasahir0) (the maintainer of this library), and Shirakawa, has been nominated as best paper at GECCO'22 ENUM track.
* **2021/03/10** ["Introduction to CMA-ES sampler"](https://medium.com/optuna/introduction-to-cma-es-sampler-ee68194c8f88) is published at Optuna Medium Blog. This article explains when and how to make the best use out of CMA-ES sampler. Please check it out!
* **2021/02/02** The paper ["Warm Starting CMA-ES for Hyperparameter Optimization"](https://arxiv.org/abs/2012.06932) written by [@nomuramasahir0](https://github.com/nomuramasahir0), the maintainer of this library, is accepted at AAAI 2021 :tada:
* **2020/07/29** Optuna's built-in CMA-ES sampler which uses this library under the hood is stabled at Optuna v2.0. Please check out the [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2).
## Installation
Supported Python versions are 3.7 or later.
```
$ pip install cmaes
```
Or you can install via [conda-forge](https://anaconda.org/conda-forge/cmaes).
```
$ conda install -c conda-forge cmaes
```
## Usage
This library provides an "ask-and-tell" style interface.
```python
import numpy as np
from cmaes import CMA
def quadratic(x1, x2):
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
optimizer = CMA(mean=np.zeros(2), sigma=1.3)
for generation in range(50):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = quadratic(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
```
And you can use this library via [Optuna](https://github.com/optuna/optuna) [2], an automatic hyperparameter optimization framework.
Optuna's built-in CMA-ES sampler which uses this library under the hood is available from [v1.3.0](https://github.com/optuna/optuna/releases/tag/v1.3.0) and stabled at [v2.0.0](https://github.com/optuna/optuna/releases/tag/v2.2.0).
See [the documentation](https://optuna.readthedocs.io/en/stable/reference/samplers/generated/optuna.samplers.CmaEsSampler.html) or [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2) for more details.
```python
import optuna
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
```
## CMA-ES variants
#### CMA-ES with Margin [3]
CMA-ES with Margin introduces a lower bound on the marginal probability associated with each discrete dimension so that samples can avoid being fixed to a single point.
It can be applied to mixed spaces of continuous (float) and discrete (including integer and binary).
|CMA-ES|CMA-ESwM|
|---|---|
|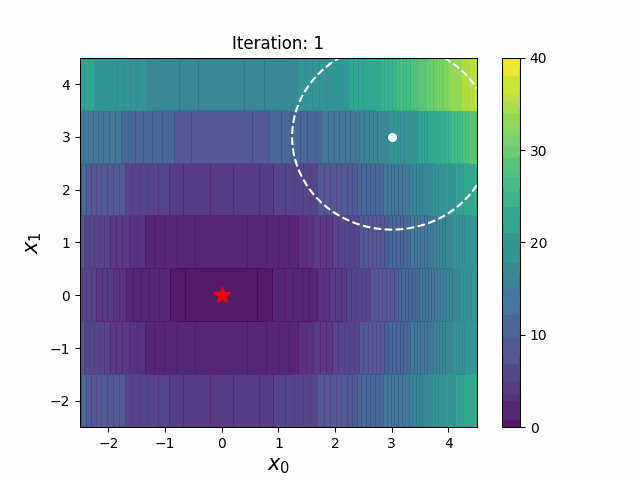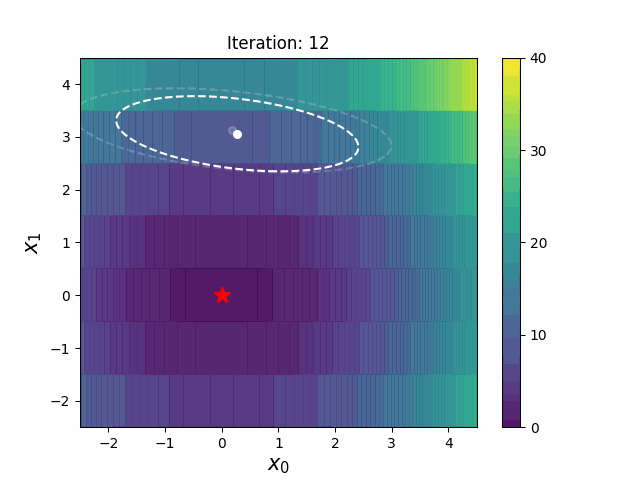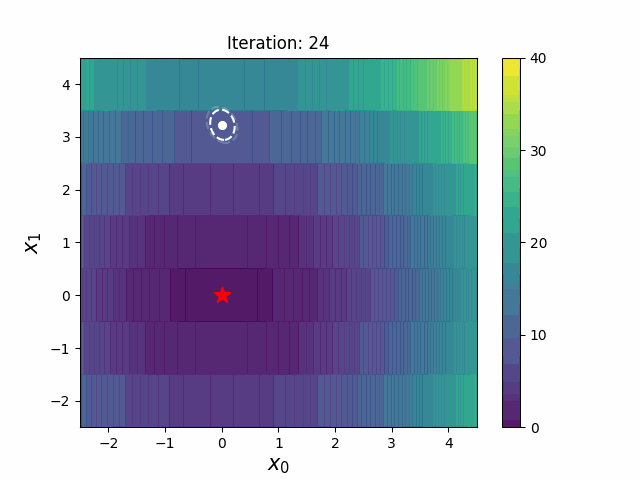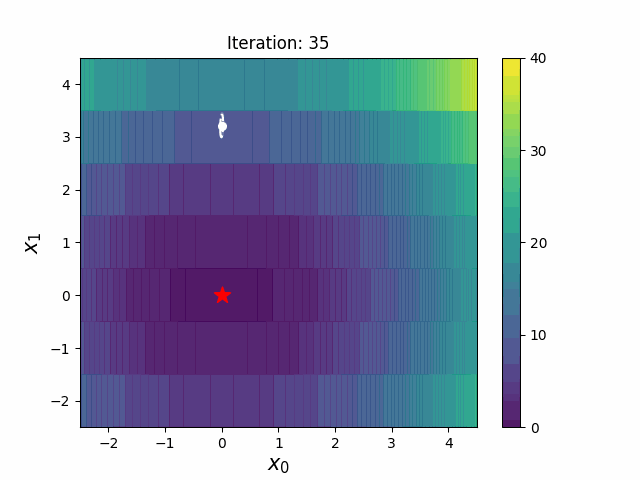|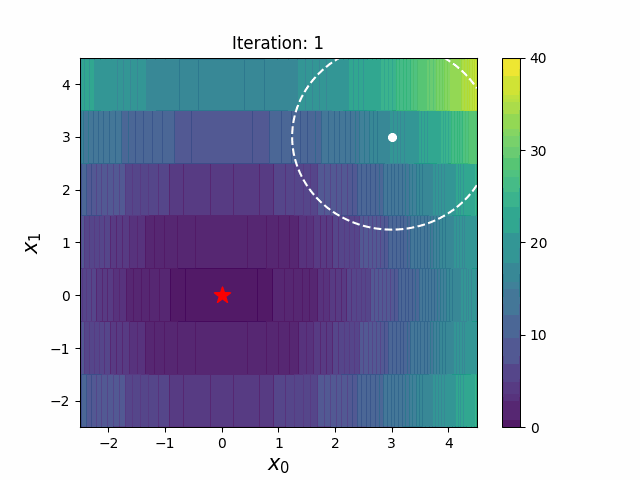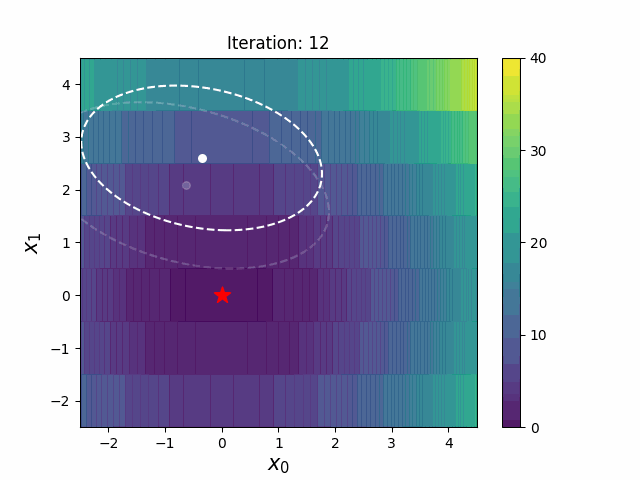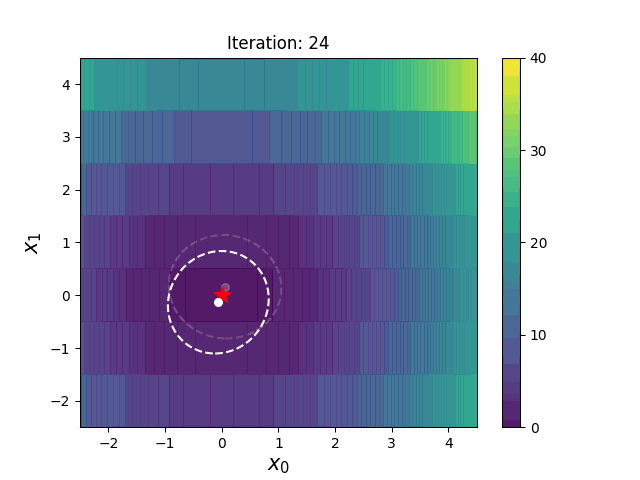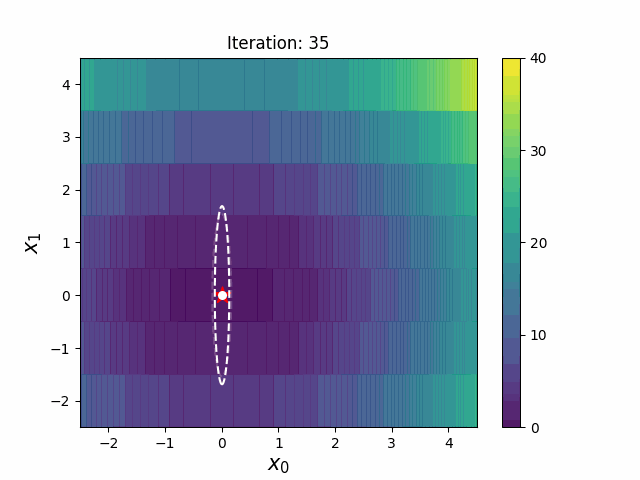|
The above figures are taken from [EvoConJP/CMA-ES_with_Margin](https://github.com/EvoConJP/CMA-ES_with_Margin).
Source code
```python
import numpy as np
from cmaes import CMAwM
def ellipsoid_onemax(x, n_zdim):
n = len(x)
n_rdim = n - n_zdim
r = 10
if len(x) < 2:
raise ValueError("dimension must be greater one")
ellipsoid = sum([(1000 ** (i / (n_rdim - 1)) * x[i]) ** 2 for i in range(n_rdim)])
onemax = n_zdim - (0.0 < x[(n - n_zdim) :]).sum()
return ellipsoid + r * onemax
def main():
binary_dim, continuous_dim = 10, 10
dim = binary_dim + continuous_dim
bounds = np.concatenate(
[
np.tile([-np.inf, np.inf], (continuous_dim, 1)),
np.tile([0, 1], (binary_dim, 1)),
]
)
steps = np.concatenate([np.zeros(continuous_dim), np.ones(binary_dim)])
optimizer = CMAwM(mean=np.zeros(dim), sigma=2.0, bounds=bounds, steps=steps)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x_for_eval, x_for_tell = optimizer.ask()
value = ellipsoid_onemax(x_for_eval, binary_dim)
evals += 1
solutions.append((x_for_tell, value))
if evals % 300 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
if __name__ == "__main__":
main()
```
Source code is also available [here](./examples/cmaes_with_margin.py).
#### Warm Starting CMA-ES [4]
Warm Starting CMA-ES is a method to transfer prior knowledge on similar HPO tasks through the initialization of CMA-ES.
Here is the result of an experiment that tuning LightGBM for Kaggle's Toxic Comment Classification Challenge data, a multilabel classification dataset.
In this benchmark, we use 10% of a full dataset as the source task, and a full dataset as the target task.
Please refer [the paper](https://arxiv.org/abs/2012.06932) and/or https://github.com/c-bata/benchmark-warm-starting-cmaes for more details of experiment settings.

Source code
```python
import numpy as np
from cmaes import CMA, get_warm_start_mgd
def source_task(x1: float, x2: float) -> float:
b = 0.4
return (x1 - b) ** 2 + (x2 - b) ** 2
def target_task(x1: float, x2: float) -> float:
b = 0.6
return (x1 - b) ** 2 + (x2 - b) ** 2
if __name__ == "__main__":
# Generate solutions from a source task
source_solutions = []
for _ in range(1000):
x = np.random.random(2)
value = source_task(x[0], x[1])
source_solutions.append((x, value))
# Estimate a promising distribution of the source task,
# then generate parameters of the multivariate gaussian distribution.
ws_mean, ws_sigma, ws_cov = get_warm_start_mgd(
source_solutions, gamma=0.1, alpha=0.1
)
optimizer = CMA(mean=ws_mean, sigma=ws_sigma, cov=ws_cov)
# Run WS-CMA-ES
print(" g f(x1,x2) x1 x2 ")
print("=== ========== ====== ======")
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = target_task(x[0], x[1])
solutions.append((x, value))
print(
f"{optimizer.generation:3d} {value:10.5f}"
f" {x[0]:6.2f} {x[1]:6.2f}"
)
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
The full source code is available [here](./examples/ws_cma_es.py).
#### Separable CMA-ES [5]
sep-CMA-ES is an algorithm which constrains the covariance matrix to be diagonal.
Due to the reduction of the number of parameters, the learning rate for the covariance matrix can be increased.
Consequently, this algorithm outperforms CMA-ES on separable functions.
Source code
```python
import numpy as np
from cmaes import SepCMA
def ellipsoid(x):
n = len(x)
if len(x) < 2:
raise ValueError("dimension must be greater one")
return sum([(1000 ** (i / (n - 1)) * x[i]) ** 2 for i in range(n)])
if __name__ == "__main__":
dim = 40
optimizer = SepCMA(mean=3 * np.ones(dim), sigma=2.0)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ellipsoid(x)
evals += 1
solutions.append((x, value))
if evals % 3000 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
Full source code is available [here](./examples/sepcma_ellipsoid_function.py).
#### IPOP-CMA-ES [6]
IPOP-CMA-ES is a method to restart CMA-ES with increasing population size like below.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
# popsize multiplied by 2 (or 3) before each restart.
popsize = optimizer.population_size * 2
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(mean=mean, sigma=sigma, population_size=popsize)
print(f"Restart CMA-ES with popsize={popsize}")
```
Full source code is available [here](./examples/ipop_cmaes.py).
#### BIPOP-CMA-ES [7]
BIPOP-CMA-ES applies two interlaced restart strategies, one with an increasing population size and one with varying small population sizes.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
n_restarts = 0 # A small restart doesn't count in the n_restarts
small_n_eval, large_n_eval = 0, 0
popsize0 = optimizer.population_size
inc_popsize = 2
# Initial run is with "normal" population size; it is
# the large population before first doubling, but its
# budget accounting is the same as in case of small
# population.
poptype = "small"
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
n_eval = optimizer.population_size * optimizer.generation
if poptype == "small":
small_n_eval += n_eval
else: # poptype == "large"
large_n_eval += n_eval
if small_n_eval < large_n_eval:
poptype = "small"
popsize_multiplier = inc_popsize ** n_restarts
popsize = math.floor(
popsize0 * popsize_multiplier ** (np.random.uniform() ** 2)
)
else:
poptype = "large"
n_restarts += 1
popsize = popsize0 * (inc_popsize ** n_restarts)
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(
mean=mean,
sigma=sigma,
bounds=bounds,
population_size=popsize,
)
print("Restart CMA-ES with popsize={} ({})".format(popsize, poptype))
```
Full source code is available [here](./examples/bipop_cmaes.py).
## Benchmark results
| [Rosenbrock function](https://www.sfu.ca/~ssurjano/rosen.html) | [Six-Hump Camel function](https://www.sfu.ca/~ssurjano/camel6.html) |
| ------------------- | ----------------------- |
|  |  |
This implementation (green) stands comparison with [pycma](https://github.com/CMA-ES/pycma) (blue).
See [benchmark](./benchmark) for details.
## Links
**Projects using cmaes:**
* [Optuna](https://github.com/optuna/optuna) : A hyperparameter optimization framework that supports CMA-ES using this library under the hood.
* (If you have a project which uses `cmaes` and want your own project to be listed here, please submit a GitHub issue.)
**Other libraries:**
I respect all libraries involved in CMA-ES.
* [pycma](https://github.com/CMA-ES/pycma) : Most famous CMA-ES implementation by Nikolaus Hansen.
* [pymoo](https://github.com/msu-coinlab/pymoo) : Multi-objective optimization in Python.
* [evojax](https://github.com/google/evojax) : EvoJAX provides a JAX-port of this library.
* [evosax](https://github.com/RobertTLange/evosax) : evosax provides JAX-based CMA-ES and sep-CMA-ES implementation, which is inspired by this library.
**References:**
* [1] [N. Hansen, The CMA Evolution Strategy: A Tutorial. arXiv:1604.00772, 2016.](https://arxiv.org/abs/1604.00772)
* [2] [T. Akiba, S. Sano, T. Yanase, T. Ohta, M. Koyama, Optuna: A Next-generation Hyperparameter Optimization Framework, KDD, 2019.](https://dl.acm.org/citation.cfm?id=3330701)
* [3] [R. Hamano, S. Saito, M. Nomura, S. Shirakawa, CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization, GECCO, 2022.](https://arxiv.org/abs/2205.13482)
* [4] [M. Nomura, S. Watanabe, Y. Akimoto, Y. Ozaki, M. Onishi, Warm Starting CMA-ES for Hyperparameter Optimization, AAAI, 2021.](https://arxiv.org/abs/2012.06932)
* [5] [R. Ros, N. Hansen, A Simple Modification in CMA-ES Achieving Linear Time and Space Complexity, PPSN, 2008.](https://hal.inria.fr/inria-00287367/document)
* [6] [A. Auger, N. Hansen, A restart CMA evolution strategy with increasing population size, CEC, 2005.](https://sci2s.ugr.es/sites/default/files/files/TematicWebSites/EAMHCO/contributionsCEC05/auger05ARCMA.pdf)
* [7] [N. Hansen, Benchmarking a BI-Population CMA-ES on the BBOB-2009 Function Testbed, GECCO Workshop, 2009.](https://hal.inria.fr/inria-00382093/document)
%package -n python3-cmaes
Summary: Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) implementation for Python 3.
Provides: python-cmaes
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-cmaes
# CMA-ES
[](./LICENSE) [](https://pypistats.org/packages/cmaes)
Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) [1] implementation.

## News
* **2022/05/13** The paper, ["CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization"](https://arxiv.org/abs/2205.13482) written by Hamano, Saito, [@nomuramasahir0](https://github.com/nomuramasahir0) (the maintainer of this library), and Shirakawa, has been nominated as best paper at GECCO'22 ENUM track.
* **2021/03/10** ["Introduction to CMA-ES sampler"](https://medium.com/optuna/introduction-to-cma-es-sampler-ee68194c8f88) is published at Optuna Medium Blog. This article explains when and how to make the best use out of CMA-ES sampler. Please check it out!
* **2021/02/02** The paper ["Warm Starting CMA-ES for Hyperparameter Optimization"](https://arxiv.org/abs/2012.06932) written by [@nomuramasahir0](https://github.com/nomuramasahir0), the maintainer of this library, is accepted at AAAI 2021 :tada:
* **2020/07/29** Optuna's built-in CMA-ES sampler which uses this library under the hood is stabled at Optuna v2.0. Please check out the [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2).
## Installation
Supported Python versions are 3.7 or later.
```
$ pip install cmaes
```
Or you can install via [conda-forge](https://anaconda.org/conda-forge/cmaes).
```
$ conda install -c conda-forge cmaes
```
## Usage
This library provides an "ask-and-tell" style interface.
```python
import numpy as np
from cmaes import CMA
def quadratic(x1, x2):
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
optimizer = CMA(mean=np.zeros(2), sigma=1.3)
for generation in range(50):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = quadratic(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
```
And you can use this library via [Optuna](https://github.com/optuna/optuna) [2], an automatic hyperparameter optimization framework.
Optuna's built-in CMA-ES sampler which uses this library under the hood is available from [v1.3.0](https://github.com/optuna/optuna/releases/tag/v1.3.0) and stabled at [v2.0.0](https://github.com/optuna/optuna/releases/tag/v2.2.0).
See [the documentation](https://optuna.readthedocs.io/en/stable/reference/samplers/generated/optuna.samplers.CmaEsSampler.html) or [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2) for more details.
```python
import optuna
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
```
## CMA-ES variants
#### CMA-ES with Margin [3]
CMA-ES with Margin introduces a lower bound on the marginal probability associated with each discrete dimension so that samples can avoid being fixed to a single point.
It can be applied to mixed spaces of continuous (float) and discrete (including integer and binary).
|CMA-ES|CMA-ESwM|
|---|---|
|||
The above figures are taken from [EvoConJP/CMA-ES_with_Margin](https://github.com/EvoConJP/CMA-ES_with_Margin).
Source code
```python
import numpy as np
from cmaes import CMAwM
def ellipsoid_onemax(x, n_zdim):
n = len(x)
n_rdim = n - n_zdim
r = 10
if len(x) < 2:
raise ValueError("dimension must be greater one")
ellipsoid = sum([(1000 ** (i / (n_rdim - 1)) * x[i]) ** 2 for i in range(n_rdim)])
onemax = n_zdim - (0.0 < x[(n - n_zdim) :]).sum()
return ellipsoid + r * onemax
def main():
binary_dim, continuous_dim = 10, 10
dim = binary_dim + continuous_dim
bounds = np.concatenate(
[
np.tile([-np.inf, np.inf], (continuous_dim, 1)),
np.tile([0, 1], (binary_dim, 1)),
]
)
steps = np.concatenate([np.zeros(continuous_dim), np.ones(binary_dim)])
optimizer = CMAwM(mean=np.zeros(dim), sigma=2.0, bounds=bounds, steps=steps)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x_for_eval, x_for_tell = optimizer.ask()
value = ellipsoid_onemax(x_for_eval, binary_dim)
evals += 1
solutions.append((x_for_tell, value))
if evals % 300 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
if __name__ == "__main__":
main()
```
Source code is also available [here](./examples/cmaes_with_margin.py).
#### Warm Starting CMA-ES [4]
Warm Starting CMA-ES is a method to transfer prior knowledge on similar HPO tasks through the initialization of CMA-ES.
Here is the result of an experiment that tuning LightGBM for Kaggle's Toxic Comment Classification Challenge data, a multilabel classification dataset.
In this benchmark, we use 10% of a full dataset as the source task, and a full dataset as the target task.
Please refer [the paper](https://arxiv.org/abs/2012.06932) and/or https://github.com/c-bata/benchmark-warm-starting-cmaes for more details of experiment settings.

Source code
```python
import numpy as np
from cmaes import CMA, get_warm_start_mgd
def source_task(x1: float, x2: float) -> float:
b = 0.4
return (x1 - b) ** 2 + (x2 - b) ** 2
def target_task(x1: float, x2: float) -> float:
b = 0.6
return (x1 - b) ** 2 + (x2 - b) ** 2
if __name__ == "__main__":
# Generate solutions from a source task
source_solutions = []
for _ in range(1000):
x = np.random.random(2)
value = source_task(x[0], x[1])
source_solutions.append((x, value))
# Estimate a promising distribution of the source task,
# then generate parameters of the multivariate gaussian distribution.
ws_mean, ws_sigma, ws_cov = get_warm_start_mgd(
source_solutions, gamma=0.1, alpha=0.1
)
optimizer = CMA(mean=ws_mean, sigma=ws_sigma, cov=ws_cov)
# Run WS-CMA-ES
print(" g f(x1,x2) x1 x2 ")
print("=== ========== ====== ======")
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = target_task(x[0], x[1])
solutions.append((x, value))
print(
f"{optimizer.generation:3d} {value:10.5f}"
f" {x[0]:6.2f} {x[1]:6.2f}"
)
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
The full source code is available [here](./examples/ws_cma_es.py).
#### Separable CMA-ES [5]
sep-CMA-ES is an algorithm which constrains the covariance matrix to be diagonal.
Due to the reduction of the number of parameters, the learning rate for the covariance matrix can be increased.
Consequently, this algorithm outperforms CMA-ES on separable functions.
Source code
```python
import numpy as np
from cmaes import SepCMA
def ellipsoid(x):
n = len(x)
if len(x) < 2:
raise ValueError("dimension must be greater one")
return sum([(1000 ** (i / (n - 1)) * x[i]) ** 2 for i in range(n)])
if __name__ == "__main__":
dim = 40
optimizer = SepCMA(mean=3 * np.ones(dim), sigma=2.0)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ellipsoid(x)
evals += 1
solutions.append((x, value))
if evals % 3000 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
Full source code is available [here](./examples/sepcma_ellipsoid_function.py).
#### IPOP-CMA-ES [6]
IPOP-CMA-ES is a method to restart CMA-ES with increasing population size like below.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
# popsize multiplied by 2 (or 3) before each restart.
popsize = optimizer.population_size * 2
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(mean=mean, sigma=sigma, population_size=popsize)
print(f"Restart CMA-ES with popsize={popsize}")
```
Full source code is available [here](./examples/ipop_cmaes.py).
#### BIPOP-CMA-ES [7]
BIPOP-CMA-ES applies two interlaced restart strategies, one with an increasing population size and one with varying small population sizes.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
n_restarts = 0 # A small restart doesn't count in the n_restarts
small_n_eval, large_n_eval = 0, 0
popsize0 = optimizer.population_size
inc_popsize = 2
# Initial run is with "normal" population size; it is
# the large population before first doubling, but its
# budget accounting is the same as in case of small
# population.
poptype = "small"
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
n_eval = optimizer.population_size * optimizer.generation
if poptype == "small":
small_n_eval += n_eval
else: # poptype == "large"
large_n_eval += n_eval
if small_n_eval < large_n_eval:
poptype = "small"
popsize_multiplier = inc_popsize ** n_restarts
popsize = math.floor(
popsize0 * popsize_multiplier ** (np.random.uniform() ** 2)
)
else:
poptype = "large"
n_restarts += 1
popsize = popsize0 * (inc_popsize ** n_restarts)
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(
mean=mean,
sigma=sigma,
bounds=bounds,
population_size=popsize,
)
print("Restart CMA-ES with popsize={} ({})".format(popsize, poptype))
```
Full source code is available [here](./examples/bipop_cmaes.py).
## Benchmark results
| [Rosenbrock function](https://www.sfu.ca/~ssurjano/rosen.html) | [Six-Hump Camel function](https://www.sfu.ca/~ssurjano/camel6.html) |
| ------------------- | ----------------------- |
|  |  |
This implementation (green) stands comparison with [pycma](https://github.com/CMA-ES/pycma) (blue).
See [benchmark](./benchmark) for details.
## Links
**Projects using cmaes:**
* [Optuna](https://github.com/optuna/optuna) : A hyperparameter optimization framework that supports CMA-ES using this library under the hood.
* (If you have a project which uses `cmaes` and want your own project to be listed here, please submit a GitHub issue.)
**Other libraries:**
I respect all libraries involved in CMA-ES.
* [pycma](https://github.com/CMA-ES/pycma) : Most famous CMA-ES implementation by Nikolaus Hansen.
* [pymoo](https://github.com/msu-coinlab/pymoo) : Multi-objective optimization in Python.
* [evojax](https://github.com/google/evojax) : EvoJAX provides a JAX-port of this library.
* [evosax](https://github.com/RobertTLange/evosax) : evosax provides JAX-based CMA-ES and sep-CMA-ES implementation, which is inspired by this library.
**References:**
* [1] [N. Hansen, The CMA Evolution Strategy: A Tutorial. arXiv:1604.00772, 2016.](https://arxiv.org/abs/1604.00772)
* [2] [T. Akiba, S. Sano, T. Yanase, T. Ohta, M. Koyama, Optuna: A Next-generation Hyperparameter Optimization Framework, KDD, 2019.](https://dl.acm.org/citation.cfm?id=3330701)
* [3] [R. Hamano, S. Saito, M. Nomura, S. Shirakawa, CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization, GECCO, 2022.](https://arxiv.org/abs/2205.13482)
* [4] [M. Nomura, S. Watanabe, Y. Akimoto, Y. Ozaki, M. Onishi, Warm Starting CMA-ES for Hyperparameter Optimization, AAAI, 2021.](https://arxiv.org/abs/2012.06932)
* [5] [R. Ros, N. Hansen, A Simple Modification in CMA-ES Achieving Linear Time and Space Complexity, PPSN, 2008.](https://hal.inria.fr/inria-00287367/document)
* [6] [A. Auger, N. Hansen, A restart CMA evolution strategy with increasing population size, CEC, 2005.](https://sci2s.ugr.es/sites/default/files/files/TematicWebSites/EAMHCO/contributionsCEC05/auger05ARCMA.pdf)
* [7] [N. Hansen, Benchmarking a BI-Population CMA-ES on the BBOB-2009 Function Testbed, GECCO Workshop, 2009.](https://hal.inria.fr/inria-00382093/document)
%package help
Summary: Development documents and examples for cmaes
Provides: python3-cmaes-doc
%description help
# CMA-ES
[](./LICENSE) [](https://pypistats.org/packages/cmaes)
Lightweight Covariance Matrix Adaptation Evolution Strategy (CMA-ES) [1] implementation.

## News
* **2022/05/13** The paper, ["CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization"](https://arxiv.org/abs/2205.13482) written by Hamano, Saito, [@nomuramasahir0](https://github.com/nomuramasahir0) (the maintainer of this library), and Shirakawa, has been nominated as best paper at GECCO'22 ENUM track.
* **2021/03/10** ["Introduction to CMA-ES sampler"](https://medium.com/optuna/introduction-to-cma-es-sampler-ee68194c8f88) is published at Optuna Medium Blog. This article explains when and how to make the best use out of CMA-ES sampler. Please check it out!
* **2021/02/02** The paper ["Warm Starting CMA-ES for Hyperparameter Optimization"](https://arxiv.org/abs/2012.06932) written by [@nomuramasahir0](https://github.com/nomuramasahir0), the maintainer of this library, is accepted at AAAI 2021 :tada:
* **2020/07/29** Optuna's built-in CMA-ES sampler which uses this library under the hood is stabled at Optuna v2.0. Please check out the [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2).
## Installation
Supported Python versions are 3.7 or later.
```
$ pip install cmaes
```
Or you can install via [conda-forge](https://anaconda.org/conda-forge/cmaes).
```
$ conda install -c conda-forge cmaes
```
## Usage
This library provides an "ask-and-tell" style interface.
```python
import numpy as np
from cmaes import CMA
def quadratic(x1, x2):
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
optimizer = CMA(mean=np.zeros(2), sigma=1.3)
for generation in range(50):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = quadratic(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
```
And you can use this library via [Optuna](https://github.com/optuna/optuna) [2], an automatic hyperparameter optimization framework.
Optuna's built-in CMA-ES sampler which uses this library under the hood is available from [v1.3.0](https://github.com/optuna/optuna/releases/tag/v1.3.0) and stabled at [v2.0.0](https://github.com/optuna/optuna/releases/tag/v2.2.0).
See [the documentation](https://optuna.readthedocs.io/en/stable/reference/samplers/generated/optuna.samplers.CmaEsSampler.html) or [v2.0 release blog](https://medium.com/optuna/optuna-v2-3165e3f1fc2) for more details.
```python
import optuna
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)
```
## CMA-ES variants
#### CMA-ES with Margin [3]
CMA-ES with Margin introduces a lower bound on the marginal probability associated with each discrete dimension so that samples can avoid being fixed to a single point.
It can be applied to mixed spaces of continuous (float) and discrete (including integer and binary).
|CMA-ES|CMA-ESwM|
|---|---|
|||
The above figures are taken from [EvoConJP/CMA-ES_with_Margin](https://github.com/EvoConJP/CMA-ES_with_Margin).
Source code
```python
import numpy as np
from cmaes import CMAwM
def ellipsoid_onemax(x, n_zdim):
n = len(x)
n_rdim = n - n_zdim
r = 10
if len(x) < 2:
raise ValueError("dimension must be greater one")
ellipsoid = sum([(1000 ** (i / (n_rdim - 1)) * x[i]) ** 2 for i in range(n_rdim)])
onemax = n_zdim - (0.0 < x[(n - n_zdim) :]).sum()
return ellipsoid + r * onemax
def main():
binary_dim, continuous_dim = 10, 10
dim = binary_dim + continuous_dim
bounds = np.concatenate(
[
np.tile([-np.inf, np.inf], (continuous_dim, 1)),
np.tile([0, 1], (binary_dim, 1)),
]
)
steps = np.concatenate([np.zeros(continuous_dim), np.ones(binary_dim)])
optimizer = CMAwM(mean=np.zeros(dim), sigma=2.0, bounds=bounds, steps=steps)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x_for_eval, x_for_tell = optimizer.ask()
value = ellipsoid_onemax(x_for_eval, binary_dim)
evals += 1
solutions.append((x_for_tell, value))
if evals % 300 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
if __name__ == "__main__":
main()
```
Source code is also available [here](./examples/cmaes_with_margin.py).
#### Warm Starting CMA-ES [4]
Warm Starting CMA-ES is a method to transfer prior knowledge on similar HPO tasks through the initialization of CMA-ES.
Here is the result of an experiment that tuning LightGBM for Kaggle's Toxic Comment Classification Challenge data, a multilabel classification dataset.
In this benchmark, we use 10% of a full dataset as the source task, and a full dataset as the target task.
Please refer [the paper](https://arxiv.org/abs/2012.06932) and/or https://github.com/c-bata/benchmark-warm-starting-cmaes for more details of experiment settings.

Source code
```python
import numpy as np
from cmaes import CMA, get_warm_start_mgd
def source_task(x1: float, x2: float) -> float:
b = 0.4
return (x1 - b) ** 2 + (x2 - b) ** 2
def target_task(x1: float, x2: float) -> float:
b = 0.6
return (x1 - b) ** 2 + (x2 - b) ** 2
if __name__ == "__main__":
# Generate solutions from a source task
source_solutions = []
for _ in range(1000):
x = np.random.random(2)
value = source_task(x[0], x[1])
source_solutions.append((x, value))
# Estimate a promising distribution of the source task,
# then generate parameters of the multivariate gaussian distribution.
ws_mean, ws_sigma, ws_cov = get_warm_start_mgd(
source_solutions, gamma=0.1, alpha=0.1
)
optimizer = CMA(mean=ws_mean, sigma=ws_sigma, cov=ws_cov)
# Run WS-CMA-ES
print(" g f(x1,x2) x1 x2 ")
print("=== ========== ====== ======")
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = target_task(x[0], x[1])
solutions.append((x, value))
print(
f"{optimizer.generation:3d} {value:10.5f}"
f" {x[0]:6.2f} {x[1]:6.2f}"
)
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
The full source code is available [here](./examples/ws_cma_es.py).
#### Separable CMA-ES [5]
sep-CMA-ES is an algorithm which constrains the covariance matrix to be diagonal.
Due to the reduction of the number of parameters, the learning rate for the covariance matrix can be increased.
Consequently, this algorithm outperforms CMA-ES on separable functions.
Source code
```python
import numpy as np
from cmaes import SepCMA
def ellipsoid(x):
n = len(x)
if len(x) < 2:
raise ValueError("dimension must be greater one")
return sum([(1000 ** (i / (n - 1)) * x[i]) ** 2 for i in range(n)])
if __name__ == "__main__":
dim = 40
optimizer = SepCMA(mean=3 * np.ones(dim), sigma=2.0)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ellipsoid(x)
evals += 1
solutions.append((x, value))
if evals % 3000 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
```
Full source code is available [here](./examples/sepcma_ellipsoid_function.py).
#### IPOP-CMA-ES [6]
IPOP-CMA-ES is a method to restart CMA-ES with increasing population size like below.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
# popsize multiplied by 2 (or 3) before each restart.
popsize = optimizer.population_size * 2
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(mean=mean, sigma=sigma, population_size=popsize)
print(f"Restart CMA-ES with popsize={popsize}")
```
Full source code is available [here](./examples/ipop_cmaes.py).
#### BIPOP-CMA-ES [7]
BIPOP-CMA-ES applies two interlaced restart strategies, one with an increasing population size and one with varying small population sizes.

Source code
```python
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
n_restarts = 0 # A small restart doesn't count in the n_restarts
small_n_eval, large_n_eval = 0, 0
popsize0 = optimizer.population_size
inc_popsize = 2
# Initial run is with "normal" population size; it is
# the large population before first doubling, but its
# budget accounting is the same as in case of small
# population.
poptype = "small"
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
n_eval = optimizer.population_size * optimizer.generation
if poptype == "small":
small_n_eval += n_eval
else: # poptype == "large"
large_n_eval += n_eval
if small_n_eval < large_n_eval:
poptype = "small"
popsize_multiplier = inc_popsize ** n_restarts
popsize = math.floor(
popsize0 * popsize_multiplier ** (np.random.uniform() ** 2)
)
else:
poptype = "large"
n_restarts += 1
popsize = popsize0 * (inc_popsize ** n_restarts)
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(
mean=mean,
sigma=sigma,
bounds=bounds,
population_size=popsize,
)
print("Restart CMA-ES with popsize={} ({})".format(popsize, poptype))
```
Full source code is available [here](./examples/bipop_cmaes.py).
## Benchmark results
| [Rosenbrock function](https://www.sfu.ca/~ssurjano/rosen.html) | [Six-Hump Camel function](https://www.sfu.ca/~ssurjano/camel6.html) |
| ------------------- | ----------------------- |
|  |  |
This implementation (green) stands comparison with [pycma](https://github.com/CMA-ES/pycma) (blue).
See [benchmark](./benchmark) for details.
## Links
**Projects using cmaes:**
* [Optuna](https://github.com/optuna/optuna) : A hyperparameter optimization framework that supports CMA-ES using this library under the hood.
* (If you have a project which uses `cmaes` and want your own project to be listed here, please submit a GitHub issue.)
**Other libraries:**
I respect all libraries involved in CMA-ES.
* [pycma](https://github.com/CMA-ES/pycma) : Most famous CMA-ES implementation by Nikolaus Hansen.
* [pymoo](https://github.com/msu-coinlab/pymoo) : Multi-objective optimization in Python.
* [evojax](https://github.com/google/evojax) : EvoJAX provides a JAX-port of this library.
* [evosax](https://github.com/RobertTLange/evosax) : evosax provides JAX-based CMA-ES and sep-CMA-ES implementation, which is inspired by this library.
**References:**
* [1] [N. Hansen, The CMA Evolution Strategy: A Tutorial. arXiv:1604.00772, 2016.](https://arxiv.org/abs/1604.00772)
* [2] [T. Akiba, S. Sano, T. Yanase, T. Ohta, M. Koyama, Optuna: A Next-generation Hyperparameter Optimization Framework, KDD, 2019.](https://dl.acm.org/citation.cfm?id=3330701)
* [3] [R. Hamano, S. Saito, M. Nomura, S. Shirakawa, CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization, GECCO, 2022.](https://arxiv.org/abs/2205.13482)
* [4] [M. Nomura, S. Watanabe, Y. Akimoto, Y. Ozaki, M. Onishi, Warm Starting CMA-ES for Hyperparameter Optimization, AAAI, 2021.](https://arxiv.org/abs/2012.06932)
* [5] [R. Ros, N. Hansen, A Simple Modification in CMA-ES Achieving Linear Time and Space Complexity, PPSN, 2008.](https://hal.inria.fr/inria-00287367/document)
* [6] [A. Auger, N. Hansen, A restart CMA evolution strategy with increasing population size, CEC, 2005.](https://sci2s.ugr.es/sites/default/files/files/TematicWebSites/EAMHCO/contributionsCEC05/auger05ARCMA.pdf)
* [7] [N. Hansen, Benchmarking a BI-Population CMA-ES on the BBOB-2009 Function Testbed, GECCO Workshop, 2009.](https://hal.inria.fr/inria-00382093/document)
%prep
%autosetup -n cmaes-0.9.1
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-cmaes -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Mon Apr 10 2023 Python_Bot - 0.9.1-1
- Package Spec generated