%global _empty_manifest_terminate_build 0
Name: python-radioactivedecay
Version: 0.4.17
Release: 1
Summary: A Python package for radioactive decay modelling that supports 1252 radionuclides, decay chains, branching, and metastable states.
License: MIT, ICRP-07, AMDC
URL: https://github.com/radioactivedecay/radioactivedecay
Source0: https://mirrors.aliyun.com/pypi/web/packages/ab/7c/23154bc6417f4e41167729055362bbe31bfddf0dfc395b91880fc88b4398/radioactivedecay-0.4.17.tar.gz
BuildArch: noarch
Requires: python3-matplotlib
Requires: python3-networkx
Requires: python3-numpy
Requires: python3-scipy
Requires: python3-setuptools
Requires: python3-sympy
Requires: python3-importlib-resources
%description
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
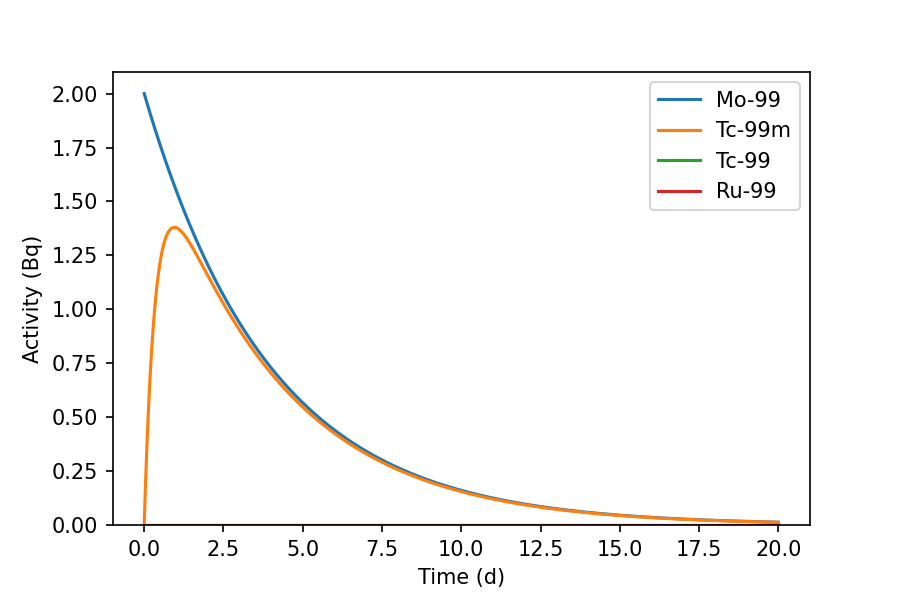 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
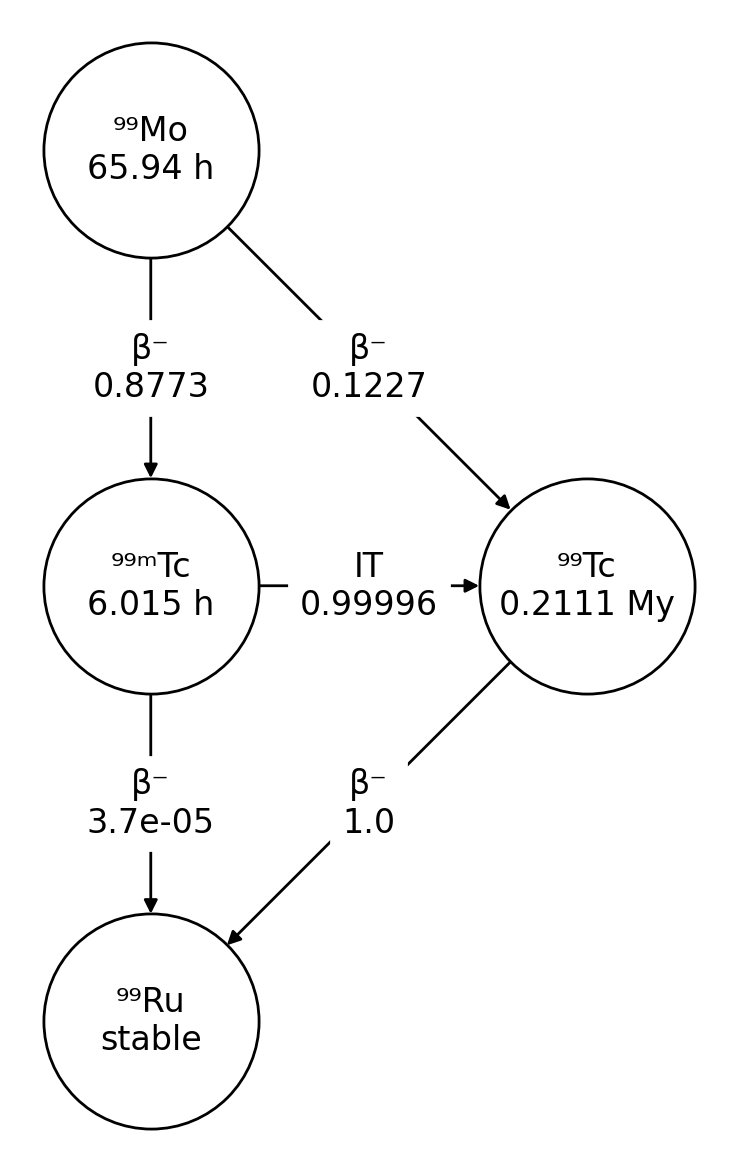 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package -n python3-radioactivedecay
Summary: A Python package for radioactive decay modelling that supports 1252 radionuclides, decay chains, branching, and metastable states.
Provides: python-radioactivedecay
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-radioactivedecay
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package -n python3-radioactivedecay
Summary: A Python package for radioactive decay modelling that supports 1252 radionuclides, decay chains, branching, and metastable states.
Provides: python-radioactivedecay
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-radioactivedecay
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package help
Summary: Development documents and examples for radioactivedecay
Provides: python3-radioactivedecay-doc
%description help
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package help
Summary: Development documents and examples for radioactivedecay
Provides: python3-radioactivedecay-doc
%description help
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%prep
%autosetup -n radioactivedecay-0.4.17
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-radioactivedecay -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri Jun 09 2023 Python_Bot - 0.4.17-1
- Package Spec generated
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%prep
%autosetup -n radioactivedecay-0.4.17
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-radioactivedecay -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri Jun 09 2023 Python_Bot - 0.4.17-1
- Package Spec generated
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package -n python3-radioactivedecay
Summary: A Python package for radioactive decay modelling that supports 1252 radionuclides, decay chains, branching, and metastable states.
Provides: python-radioactivedecay
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-radioactivedecay
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package -n python3-radioactivedecay
Summary: A Python package for radioactive decay modelling that supports 1252 radionuclides, decay chains, branching, and metastable states.
Provides: python-radioactivedecay
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-radioactivedecay
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package help
Summary: Development documents and examples for radioactivedecay
Provides: python3-radioactivedecay-doc
%description help
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%package help
Summary: Development documents and examples for radioactivedecay
Provides: python3-radioactivedecay-doc
%description help
 ***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
***
[](https://pypi.org/project/radioactivedecay/)
[](https://anaconda.org/conda-forge/radioactivedecay)
[](https://pypi.org/project/radioactivedecay/)
[](https://radioactivedecay.github.io/)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/1_tests.yml)
[](https://codecov.io/gh/radioactivedecay/radioactivedecay)
[](https://github.com/radioactivedecay/radioactivedecay/actions/workflows/3_code_formatting.yml)
[](https://doi.org/10.21105/joss.03318)
[](https://pepy.tech/project/radioactivedecay)
``radioactivedecay`` is a Python package for radioactive decay calculations.
It supports decay chains of radionuclides, metastable states and branching
decays. By default it uses the decay data from ICRP Publication 107, which
contains 1252 radionuclides of 97 elements, and atomic mass data from the
Atomic Mass Data Center.
The code solves the radioactive decay differential equations analytically using
NumPy and SciPy linear algebra routines. There is also a high numerical
precision calculation mode employing SymPy routines. This gives more accurate
results for decay chains containing radionuclides with orders of magnitude
differences between the half-lives.
This is free-to-use open source software. It was created for engineers,
technicians and researchers who work with radioactivity, and for
educational use.
- **Full Documentation**:
[https://radioactivedecay.github.io/](https://radioactivedecay.github.io/)
## Installation
``radioactivedecay`` requires Python 3.6+. Install ``radioactivedecay`` from
the [Python Package Index](https://pypi.org/project/radioactivedecay/) using
``pip``:
```console
$ pip install radioactivedecay
```
or from [conda-forge](https://anaconda.org/conda-forge/radioactivedecay):
```console
$ conda install -c conda-forge radioactivedecay
```
Either command will attempt to install the dependencies (Matplotlib, NetworkX,
NumPy, SciPy, Setuptools & SymPy) if they are not already present in the
environment.
## Usage
### Decay calculations
Create an ``Inventory`` of radionuclides and decay it as follows:
```pycon
>>> import radioactivedecay as rd
>>> Mo99_t0 = rd.Inventory({'Mo-99': 2.0}, 'Bq')
>>> Mo99_t1 = Mo99_t0.decay(20.0, 'h')
>>> Mo99_t1.activities('Bq')
{'Mo-99': 1.6207863893776937, 'Ru-99': 0.0,
'Tc-99': 9.05304236308454e-09, 'Tc-99m': 1.3719829376710406}
```
An ``Inventory`` of 2.0 Bq of Mo-99 was decayed for 20 hours, producing the
radioactive progeny Tc-99m and Tc-99, and the stable nuclide Ru-99.
We supplied ``'h'`` as an argument to ``decay()`` to specify the decay time
period had units of hours. Supported time units include ``'μs'``, ``'ms'``,
``'s'``, ``'m'``, ``'h'``, ``'d'``, ``'y'`` etc. Note seconds (``'s'``) is the
default if no unit is supplied to ``decay()``.
Use `cumulative_decays()` to calculate the total number of atoms of each
radionuclide that decay over the decay time period:
```pycon
>>> Mo99_t0.cumulative_decays(20.0, 'h')
{'Mo-99': 129870.3165339939, 'Tc-99m': 71074.31925850797,
'Tc-99': 0.0002724635511147602}
```
Radionuclides can be specified in four equivalent ways in ``radioactivedecay``:
three variations of nuclide strings or by
[canonical ids](https://pyne.io/usersguide/nucname.html). For example, the
following are equivalent ways of specifying 222Rn and
192nIr:
* ``'Rn-222'``, ``'Rn222'``, ``'222Rn'``, ``862220000``,
* ``'Ir-192n'``, ``'Ir192n'``, ``'192nIr'``, ``771920002``.
Inventories can be created by supplying activity (``'Bq'``, ``'Ci'``,
``'dpm'``...), mass (``'g'``, ``'kg'``...), mole (``'mol'``, ``'kmol'``...)
units, or numbers of nuclei (``'num'``) to the ``Inventory()`` constructor. Use
the methods ``activities()``, ``masses()``, ``moles()``, ``numbers()``,
``activity_fractions()``, ``mass_fractions()`` and ``mole_fractions()`` to
obtain the contents of the inventory in different formats:
```pycon
>>> H3_t0 = rd.Inventory({'H-3': 3.0}, 'g')
>>> H3_t1 = H3_t0.decay(12.32, 'y')
>>> H3_t1.masses('g')
{'H-3': 1.5, 'He-3': 1.4999900734297729}
>>> H3_t1.mass_fractions()
{'H-3': 0.5000016544338455, 'He-3': 0.4999983455661545}
>>> C14_t0 = rd.Inventory({'C-14': 3.2E24}, 'num')
>>> C14_t1 = C14_t0.decay(3000, 'y')
>>> C14_t1.moles('mol')
{'C-14': 3.6894551567795797, 'N-14': 1.6242698581767292}
>>> C14_t1.mole_fractions()
{'C-14': 0.6943255713073281, 'N-14': 0.3056744286926719}
```
### Plotting decay graphs
Use the ``plot()`` method to graph of the decay of an inventory over time:
```pycon
>>> Mo99_t0.plot(20, 'd', yunits='Bq')
```
 The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
The graph shows the decay of Mo-99 over 20 days, leading to the ingrowth of
Tc-99m and a trace quantity of Tc-99. The activity of Ru-99 is strictly zero as
it is the stable nuclide at the end of the decay chain. Graphs are drawn using
Matplotlib.
### Fetching decay data
The ``Nuclide`` class can be used to fetch decay information for
individual radionuclides, e.g. for Rn-222:
```pycon
>>> nuc = rd.Nuclide('Rn-222')
>>> nuc.half_life('s')
330350.4
>>> nuc.half_life('readable')
'3.8235 d'
>>> nuc.progeny()
['Po-218']
>>> nuc.branching_fractions()
[1.0]
>>> nuc.decay_modes()
['α']
>>> nuc.Z # proton number
86
>>> nuc.A # nucleon number
222
>>> nuc.atomic_mass # atomic mass in g/mol
222.01757601699998
```
There are similar inventory methods for fetching decay data:
```pycon
>>> Mo99_t1.half_lives('readable')
{'Mo-99': '65.94 h', 'Ru-99': 'stable', 'Tc-99': '0.2111 My', 'Tc-99m': '6.015 h'}
>>> Mo99_t1.progeny()
{'Mo-99': ['Tc-99m', 'Tc-99'], 'Ru-99': [], 'Tc-99': ['Ru-99'], 'Tc-99m': ['Tc-99', 'Ru-99']}
>>> Mo99_t1.branching_fractions()
{'Mo-99': [0.8773, 0.1227], 'Ru-99': [], 'Tc-99': [1.0], 'Tc-99m': [0.99996, 3.7e-05]}
>>> Mo99_t1.decay_modes()
{'Mo-99': ['β-', 'β-'], 'Ru-99': [], 'Tc-99': ['β-'], 'Tc-99m': ['IT', 'β-']}
```
### Decay chain diagrams
The ``Nuclide`` class includes a `plot()` method for drawing decay chain
diagrams:
```pycon
>>> nuc = rd.Nuclide('Mo-99')
>>> nuc.plot()
```
 These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%prep
%autosetup -n radioactivedecay-0.4.17
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-radioactivedecay -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri Jun 09 2023 Python_Bot
These diagrams are drawn using NetworkX and Matplotlib.
### High numerical precision decay calculations
``radioactivedecay`` includes an ``InventoryHP`` class for high numerical
precision calculations. This class can give more reliable decay calculation
results for chains containing long- and short-lived radionuclides:
```pycon
>>> U238_t0 = rd.InventoryHP({'U-238': 1.0})
>>> U238_t1 = U238_t0.decay(10.0, 'd')
>>> U238_t1.activities()
{'At-218': 1.4511675857141352e-25,
'Bi-210': 1.8093327888942224e-26,
'Bi-214': 7.09819414496093e-22,
'Hg-206': 1.9873081129046843e-33,
'Pa-234': 0.00038581180879502017,
'Pa-234m': 0.24992285949158477,
'Pb-206': 0.0,
'Pb-210': 1.0508864357335218e-25,
'Pb-214': 7.163682655782086e-22,
'Po-210': 1.171277829871092e-28,
'Po-214': 7.096704966148592e-22,
'Po-218': 7.255923469955255e-22,
'Ra-226': 2.6127168262000313e-21,
'Rn-218': 1.4511671865210924e-28,
'Rn-222': 7.266530698712501e-22,
'Th-230': 8.690585458641225e-16,
'Th-234': 0.2499481473619856,
'Tl-206': 2.579902288672889e-32,
'Tl-210': 1.4897029111914831e-25,
'U-234': 1.0119788393651999e-08,
'U-238': 0.9999999999957525}
```
## How radioactivedecay works
``radioactivedecay`` calculates an analytical solution to the radioactive decay
differential equations using linear algebra operations. It implements the
method described in this paper:
[M Amaku, PR Pascholati & VR Vanin, Comp. Phys. Comm. 181, 21-23
(2010)](https://doi.org/10.1016/j.cpc.2009.08.011). See the
[theory docpage](https://radioactivedecay.github.io/theory.html) for more
details.
It uses NumPy and SciPy routines for standard decay calculations
(double-precision floating-point operations), and SymPy for arbitrary numerical
precision calculations.
By default ``radioactivedecay`` uses decay data from
[ICRP Publication 107
(2008)](https://journals.sagepub.com/doi/pdf/10.1177/ANIB_38_3) and atomic mass
data from the [Atomic Mass Data Center](https://www-nds.iaea.org/amdc/)
(AMDC - AME2020 and Nubase2020 evaluations).
The [datasets repo](https://github.com/radioactivedecay/datasets) contains
Jupyter Notebooks for creating decay datasets that can be used by
``radioactivedecay``, e.g. [ICRP
107](https://github.com/radioactivedecay/datasets/blob/main/icrp107_ame2020_nubase2020/icrp107_dataset.ipynb).
The [comparisons repo](https://github.com/radioactivedecay/comparisons)
contains some checks of ``radioactivedecay`` against
[PyNE](https://github.com/radioactivedecay/comparisons/blob/main/pyne/rd_pyne_truncated_compare.ipynb)
and [Radiological
Toolbox](https://github.com/radioactivedecay/comparisons/blob/main/radiological_toolbox/radiological_toolbox_compare.ipynb).
## Tests
From the base directory run:
```console
$ python -m unittest discover
```
## License
``radioactivedecay`` is open source software released under the MIT License.
See [LICENSE](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE)
file for details.
The default decay data used by ``radioactivedecay`` (ICRP-107) is copyright
2008 A. Endo and K.F. Eckerman and distributed under a separate
[license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.ICRP-07).
The default atomic mass data is from AMDC
([license](https://github.com/radioactivedecay/radioactivedecay/blob/main/LICENSE.AMDC)).
## Citation
If you find this package useful for your research, please consider citing the
paper on ``radioactivedecay`` published in the
[Journal of Open Source Software](https://doi.org/10.21105/joss.03318):
> Alex Malins & Thom Lemoine, *radioactivedecay: A Python package for radioactive decay
calculations*. Journal of Open Source Software, **7** (71), 3318 (2022). DOI:
[10.21105/joss.03318](https://doi.org/10.21105/joss.03318).
## Contributing
Contributors are welcome to fix bugs, add new features or make feature
requests. Please open an Issue, Pull Request or new Discussions thread at
[GitHub repository](https://github.com/radioactivedecay/radioactivedecay).
Please read the
[contribution guidelines](https://github.com/radioactivedecay/radioactivedecay/blob/main/CONTRIBUTING.md).
%prep
%autosetup -n radioactivedecay-0.4.17
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-radioactivedecay -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri Jun 09 2023 Python_Bot