%global _empty_manifest_terminate_build 0
Name: python-amici
Version: 0.16.1
Release: 1
Summary: Advanced multi-language Interface to CVODES and IDAS
License: BSD 3-Clause License
URL: https://github.com/AMICI-dev/AMICI
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/a3/01/dda1b86559667af98c446493de359c119dc776128c43310d14a9200d0d56/amici-0.16.1.tar.gz
BuildArch: noarch
%description
 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status






 ## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
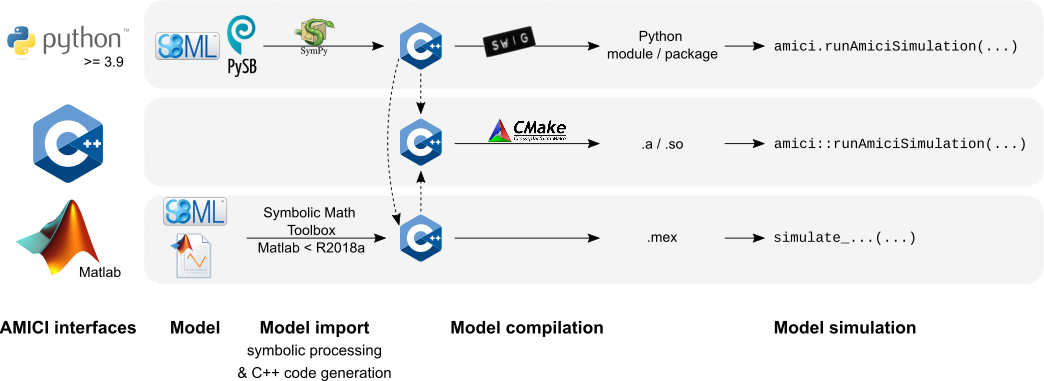
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:
## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.

## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:

%package -n python3-amici
Summary: Advanced multi-language Interface to CVODES and IDAS
Provides: python-amici
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-amici
 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status






 ## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.

## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:
## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.

## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:

%package help
Summary: Development documents and examples for amici
Provides: python3-amici-doc
%description help
 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status






 ## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.

## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:
## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.

## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/develop/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/develop/matlab_installation.html).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/master/documentation/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/develop?labpath=documentation%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/master/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/develop/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[documentation/gfx/](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/master/documentation/gfx/LICENSE.md)
license:

%prep
%autosetup -n amici-0.16.1
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-amici -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri May 05 2023 Python_Bot - 0.16.1-1
- Package Spec generated

 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status

 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status

 ## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
In contrast to the (no longer maintained)
[sundialsTB](https://computing.llnl.gov/projects/sundials/sundials-software)
Matlab interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
