%global _empty_manifest_terminate_build 0
Name: python-spacemake
Version: 0.7.2
Release: 1
Summary: A bioinformatic pipeline for the analysis of spatial transcriptomic data
License: GPL
URL: https://github.com/rajewsky-lab/spacemake
Source0: https://mirrors.aliyun.com/pypi/web/packages/fb/22/98eb4954a6ca3e55904ae0cac75d64a75b93aec06096a496a909bcc95269/spacemake-0.7.2.tar.gz
BuildArch: noarch
%description


 # Spacemake: processing and analysis of large-scale spatial transcriptomics data
# Spacemake: processing and analysis of large-scale spatial transcriptomics data
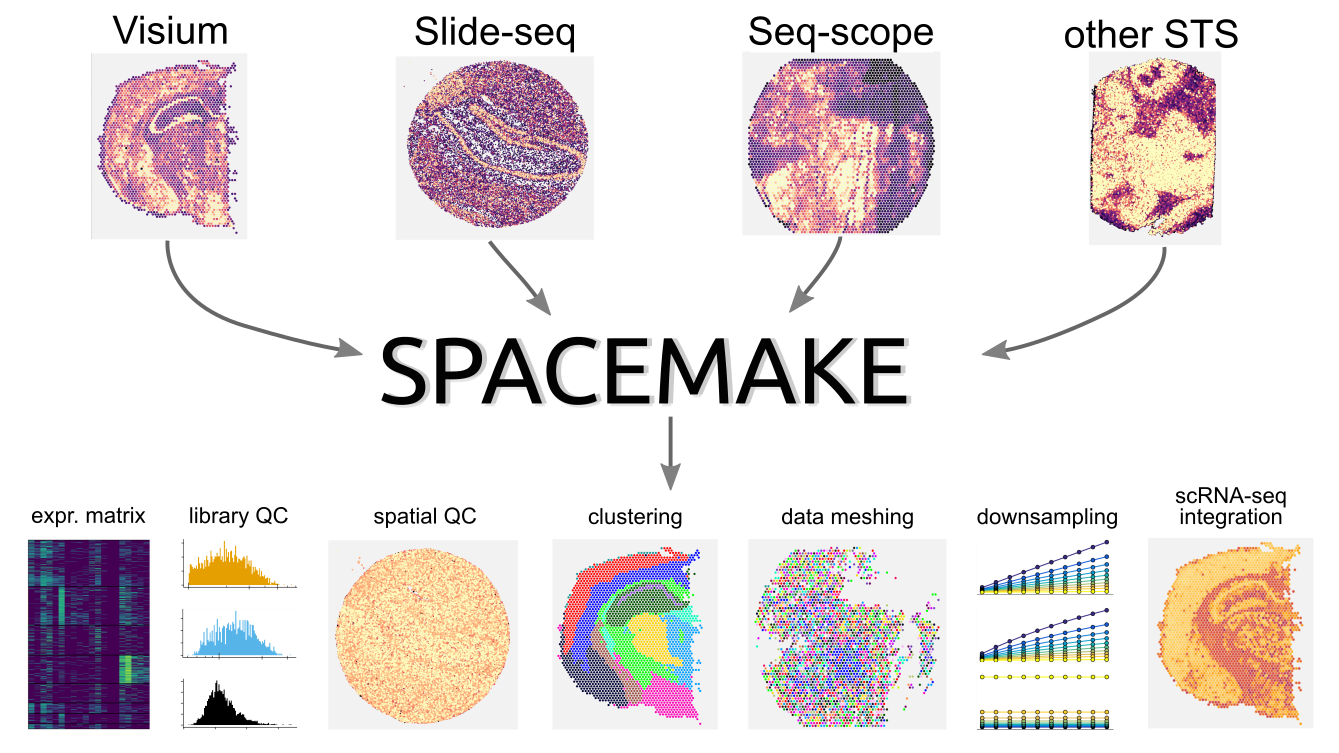 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package -n python3-spacemake
Summary: A bioinformatic pipeline for the analysis of spatial transcriptomic data
Provides: python-spacemake
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-spacemake
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package -n python3-spacemake
Summary: A bioinformatic pipeline for the analysis of spatial transcriptomic data
Provides: python-spacemake
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-spacemake


 # Spacemake: processing and analysis of large-scale spatial transcriptomics data
# Spacemake: processing and analysis of large-scale spatial transcriptomics data
 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package help
Summary: Development documents and examples for spacemake
Provides: python3-spacemake-doc
%description help
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package help
Summary: Development documents and examples for spacemake
Provides: python3-spacemake-doc
%description help


 # Spacemake: processing and analysis of large-scale spatial transcriptomics data
# Spacemake: processing and analysis of large-scale spatial transcriptomics data
 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%prep
%autosetup -n spacemake-0.7.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-spacemake -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot - 0.7.2-1
- Package Spec generated
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%prep
%autosetup -n spacemake-0.7.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-spacemake -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot - 0.7.2-1
- Package Spec generated

 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package -n python3-spacemake
Summary: A bioinformatic pipeline for the analysis of spatial transcriptomic data
Provides: python-spacemake
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-spacemake
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package -n python3-spacemake
Summary: A bioinformatic pipeline for the analysis of spatial transcriptomic data
Provides: python-spacemake
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-spacemake

 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package help
Summary: Development documents and examples for spacemake
Provides: python3-spacemake-doc
%description help
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%package help
Summary: Development documents and examples for spacemake
Provides: python3-spacemake-doc
%description help

 Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%prep
%autosetup -n spacemake-0.7.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-spacemake -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot
Spacemake is a modular, robust, and scalable spatial transcriptomics pipeline built in `Snakemake` and `Python`. Spacemake is designed to handle all major spatial transcriptomics datasets and can be readily configured for other technologies. It can process and analyze several samples in parallel, even if they stem from different experimental methods. Spacemake's unified framework enables reproducible data processing from raw sequencing data to automatically generated downstream analysis reports. Spacemake is built with a modular design and offers additional functionality such as sample merging, saturation analysis, and analysis of long reads as separate modules.
If you find Spacemake useful in your work, consider citing it:
```
Spacemake: processing and analysis of large-scale spatial transcriptomics data
Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
GigaScience, Volume 11, 2022, giac064
```
Documentation can be found [here](https://spacemake.readthedocs.io/en/latest/).
%prep
%autosetup -n spacemake-0.7.2
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-spacemake -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot