%global _empty_manifest_terminate_build 0
Name: python-jupyterlab-mol-visualizer
Version: 0.3.0
Release: 1
Summary: A JupyterLab launcher extension to view the molecular orbitals.
License: BSD 3-Clause License Copyright (c) 2023, Dou Du All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met: 1. Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. 2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. 3. Neither the name of the copyright holder nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
URL: https://github.com/osscar-org/jupyterlab-mol-visualizer
Source0: https://mirrors.aliyun.com/pypi/web/packages/9f/d0/6c8a0bae46916ab3142ea6a87f6299194bc0ae4eaad0d68073048492f8a1/jupyterlab_mol_visualizer-0.3.0.tar.gz
BuildArch: noarch
%description
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.
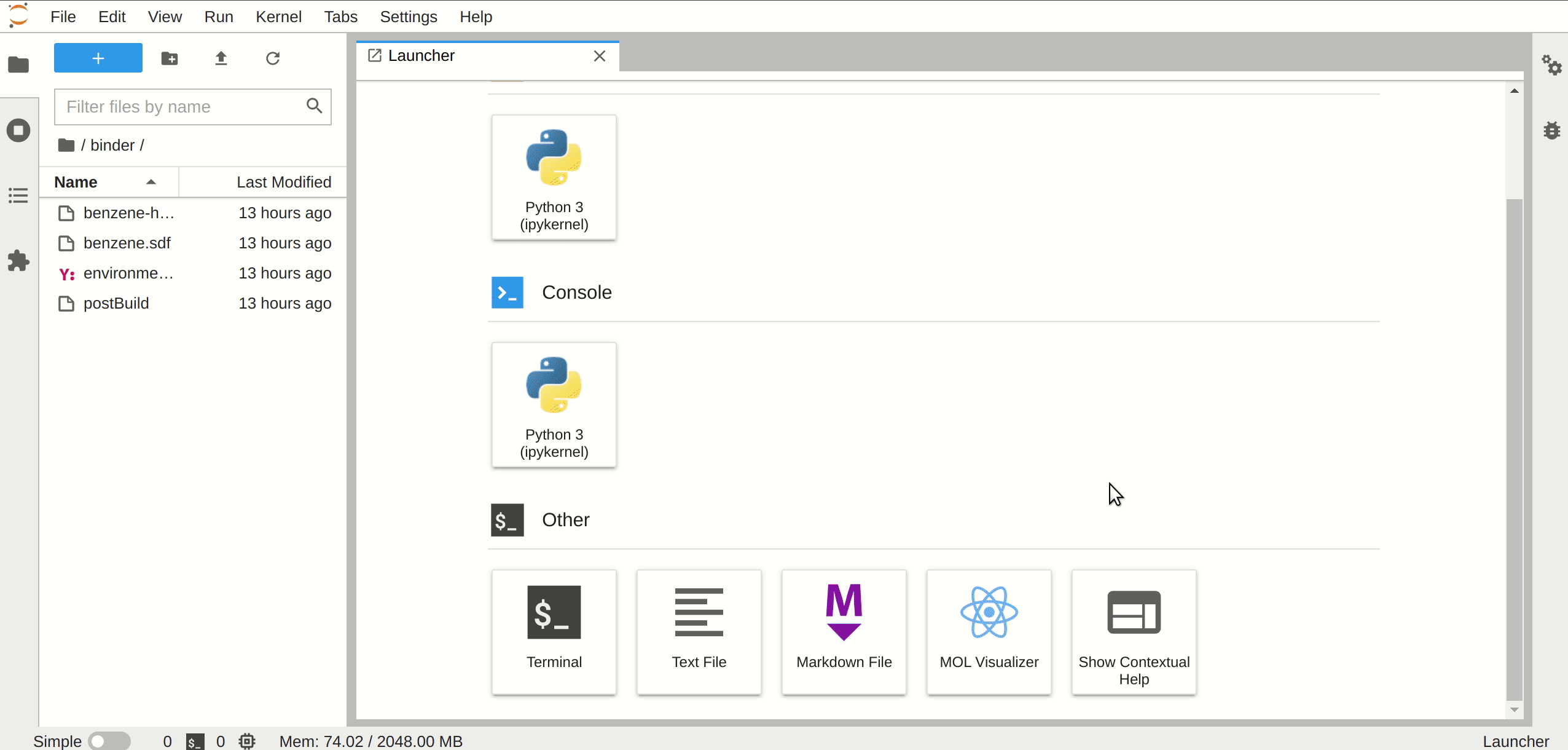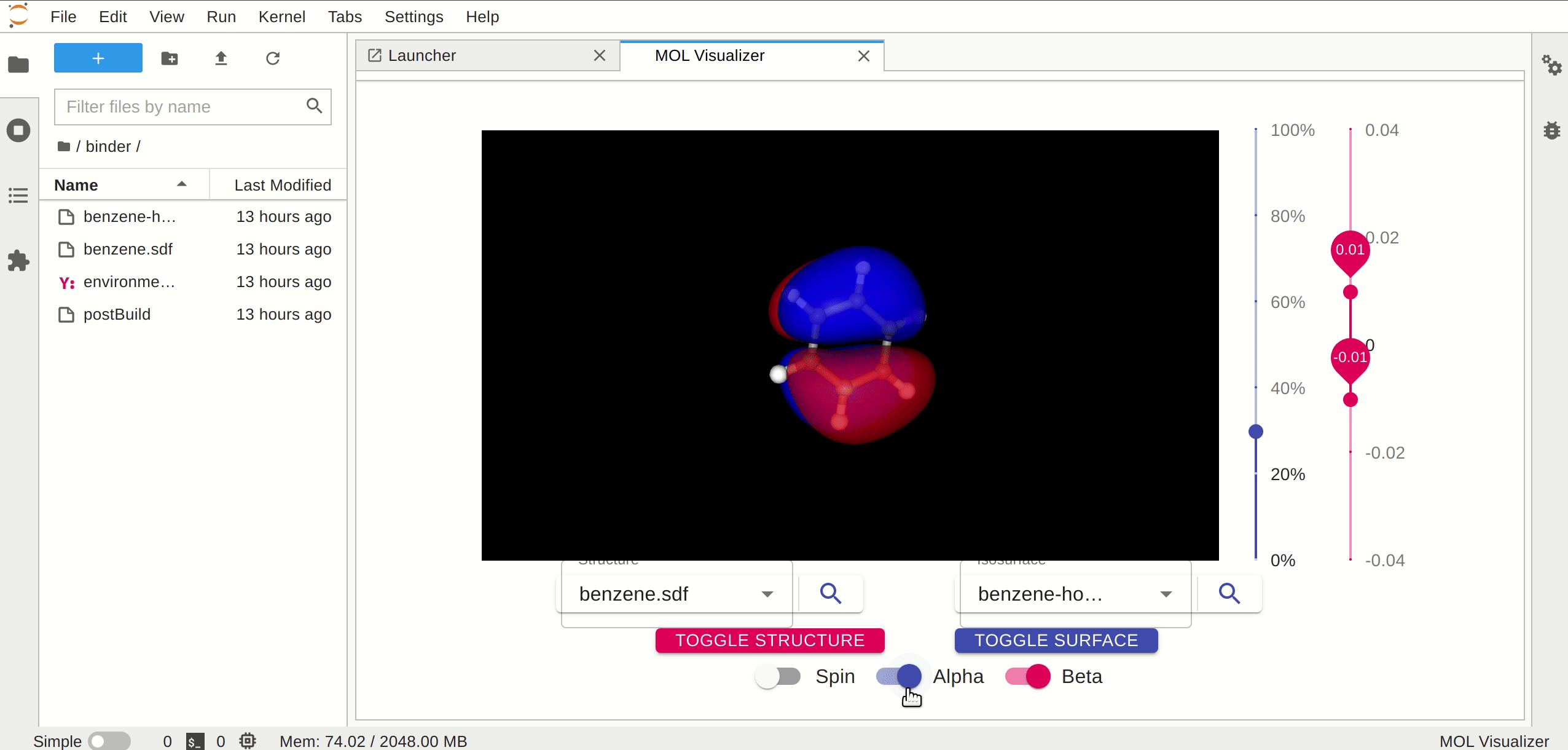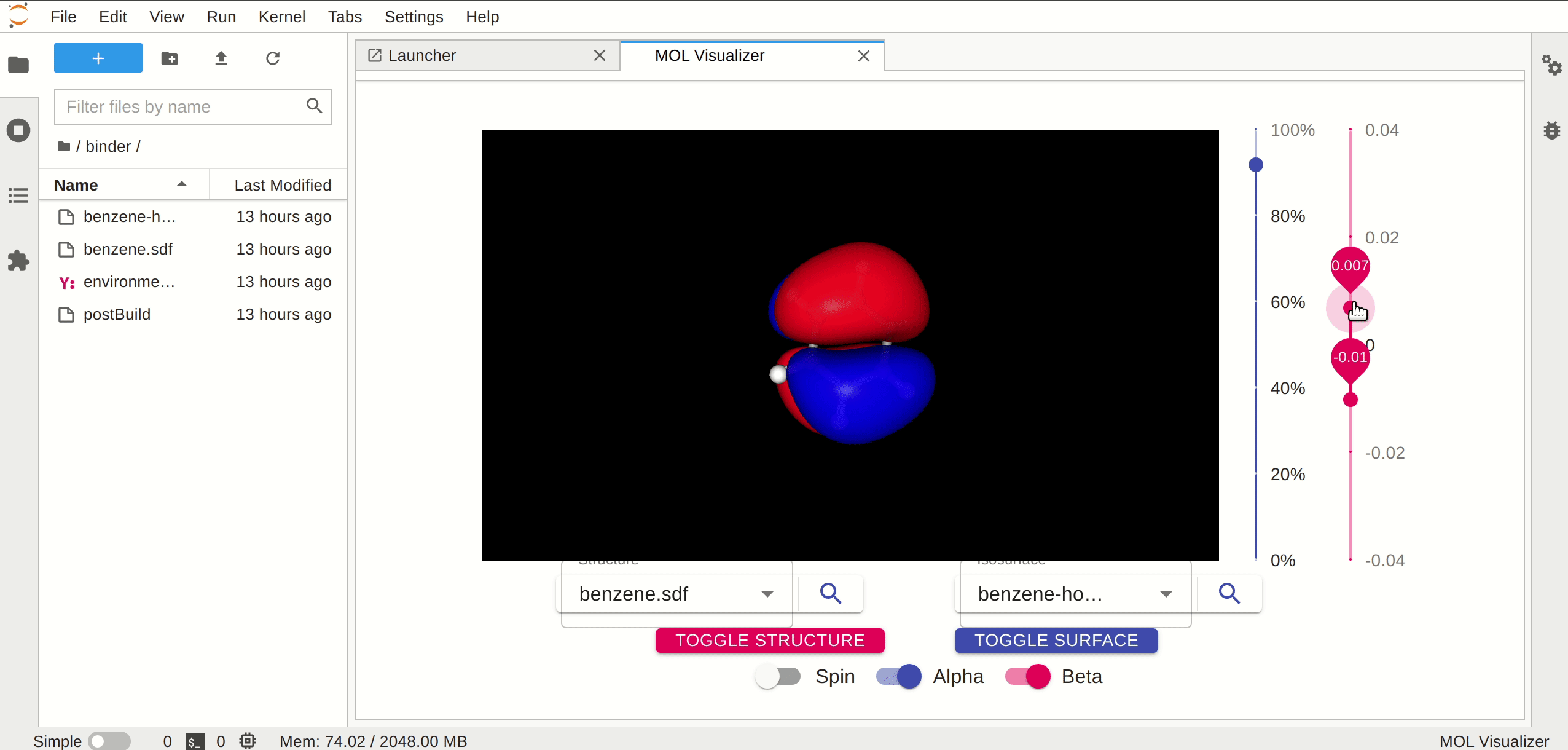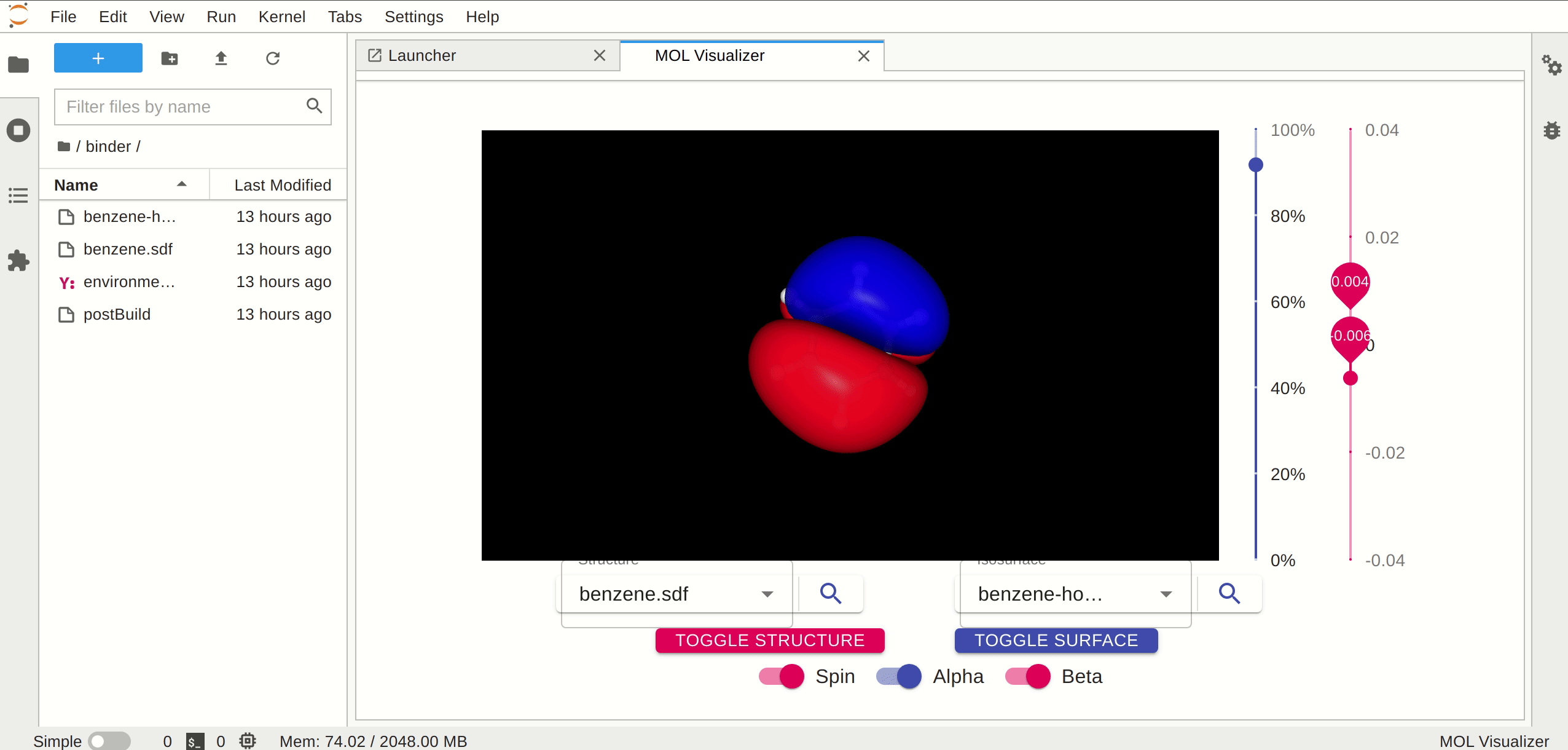
## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
 %package -n python3-jupyterlab-mol-visualizer
Summary: A JupyterLab launcher extension to view the molecular orbitals.
Provides: python-jupyterlab-mol-visualizer
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-jupyterlab-mol-visualizer
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
%package -n python3-jupyterlab-mol-visualizer
Summary: A JupyterLab launcher extension to view the molecular orbitals.
Provides: python-jupyterlab-mol-visualizer
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-jupyterlab-mol-visualizer
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
 %package help
Summary: Development documents and examples for jupyterlab-mol-visualizer
Provides: python3-jupyterlab-mol-visualizer-doc
%description help
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
%package help
Summary: Development documents and examples for jupyterlab-mol-visualizer
Provides: python3-jupyterlab-mol-visualizer-doc
%description help
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
 %prep
%autosetup -n jupyterlab_mol_visualizer-0.3.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-jupyterlab-mol-visualizer -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot - 0.3.0-1
- Package Spec generated
%prep
%autosetup -n jupyterlab_mol_visualizer-0.3.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-jupyterlab-mol-visualizer -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot - 0.3.0-1
- Package Spec generated
 %package -n python3-jupyterlab-mol-visualizer
Summary: A JupyterLab launcher extension to view the molecular orbitals.
Provides: python-jupyterlab-mol-visualizer
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-jupyterlab-mol-visualizer
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
%package -n python3-jupyterlab-mol-visualizer
Summary: A JupyterLab launcher extension to view the molecular orbitals.
Provides: python-jupyterlab-mol-visualizer
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-jupyterlab-mol-visualizer
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
 %package help
Summary: Development documents and examples for jupyterlab-mol-visualizer
Provides: python3-jupyterlab-mol-visualizer-doc
%description help
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
%package help
Summary: Development documents and examples for jupyterlab-mol-visualizer
Provides: python3-jupyterlab-mol-visualizer-doc
%description help
# **jupyterlab-mol-visualizer**: A JupyterLab Launcher to Visualize Molecular Orbital and Structure

[](https://badge.fury.io/py/jupyterlab-mol-visualizer)
[](https://badge.fury.io/js/jupyterlab-mol-visualizer)
## Try it with Binder
[](https://mybinder.org/v2/gh/osscar-org/jupyterlab-mol-visualizer/master?urlpath=lab)
A JupyterLab launcher extension to view the molecular orbitals.

## Requirements
- JupyterLab >= 3.0
## Install
To install the extension, execute:
```bash
pip install jupyterlab-mol-visualizer
```
or
```bash
jupyter labextension install jupyterlab-mol-visualizer
```
## Uninstall
To remove the extension, execute:
```bash
pip uninstall jupyterlab-mol-visualizer
```
## Contributing
### Development install
Note: You will need NodeJS to build the extension package.
The `jlpm` command is JupyterLab's pinned version of
[yarn](https://yarnpkg.com/) that is installed with JupyterLab. You may use
`yarn` or `npm` in lieu of `jlpm` below.
```bash
# Clone the repo to your local environment
# Change directory to the jupyterlab_mol_visualizer directory
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild extension Typescript source after making changes
jlpm run build
```
You can watch the source directory and run JupyterLab at the same time in different terminals to watch for changes in the extension's source and automatically rebuild the extension.
```bash
# Watch the source directory in one terminal, automatically rebuilding when needed
jlpm run watch
# Run JupyterLab in another terminal
jupyter lab
```
With the watch command running, every saved change will immediately be built locally and available in your running JupyterLab. Refresh JupyterLab to load the change in your browser (you may need to wait several seconds for the extension to be rebuilt).
By default, the `jlpm run build` command generates the source maps for this extension to make it easier to debug using the browser dev tools. To also generate source maps for the JupyterLab core extensions, you can run the following command:
```bash
jupyter lab build --minimize=False
```
### Development uninstall
```bash
pip uninstall jupyterlab_mol_visualizer
```
In development mode, you will also need to remove the symlink created by `jupyter labextension develop`
command. To find its location, you can run `jupyter labextension list` to figure out where the `labextensions`
folder is located. Then you can remove the symlink named `jupyterlab-mol-visualizer` within that folder.
## Acknowledgements
We acknowledge support from the EPFL Open Science Fund via the [OSSCAR](http://www.osscar.org) project.
 %prep
%autosetup -n jupyterlab_mol_visualizer-0.3.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-jupyterlab-mol-visualizer -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot
%prep
%autosetup -n jupyterlab_mol_visualizer-0.3.0
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "\"/%h/%f\"\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "\"/%h/%f.gz\"\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-jupyterlab-mol-visualizer -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue Jun 20 2023 Python_Bot