blob: 532a1b55bffdeeb31782b4219a54dd63621c7eff (
plain)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
|
%global _empty_manifest_terminate_build 0
Name: python-ATACFragQC
Version: 0.4.6
Release: 1
Summary: Fragment Quality Control for ATAC-seq
License: MIT
URL: https://github.com/0CBH0/ATACFragQC
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/83/b4/d670c2a7fdd5d9e594ddea6f0646bca46cddfda7a09624befef8ea4a3c35/ATACFragQC-0.4.6.tar.gz
BuildArch: noarch
%description
# ATACFragQC
The Python toolkits designed to control the fragment quality of Bulk/SingCell ATAC-seq.
## Installation
~~~
python3 -m pip install --upgrade ATACFragQC
~~~
## Usage
~~~
# Basic usage
ATACFragQC [options] -i <input.bam> -r <reference.gtf>
# For more information
ATACFragQC -h
~~~
## Features
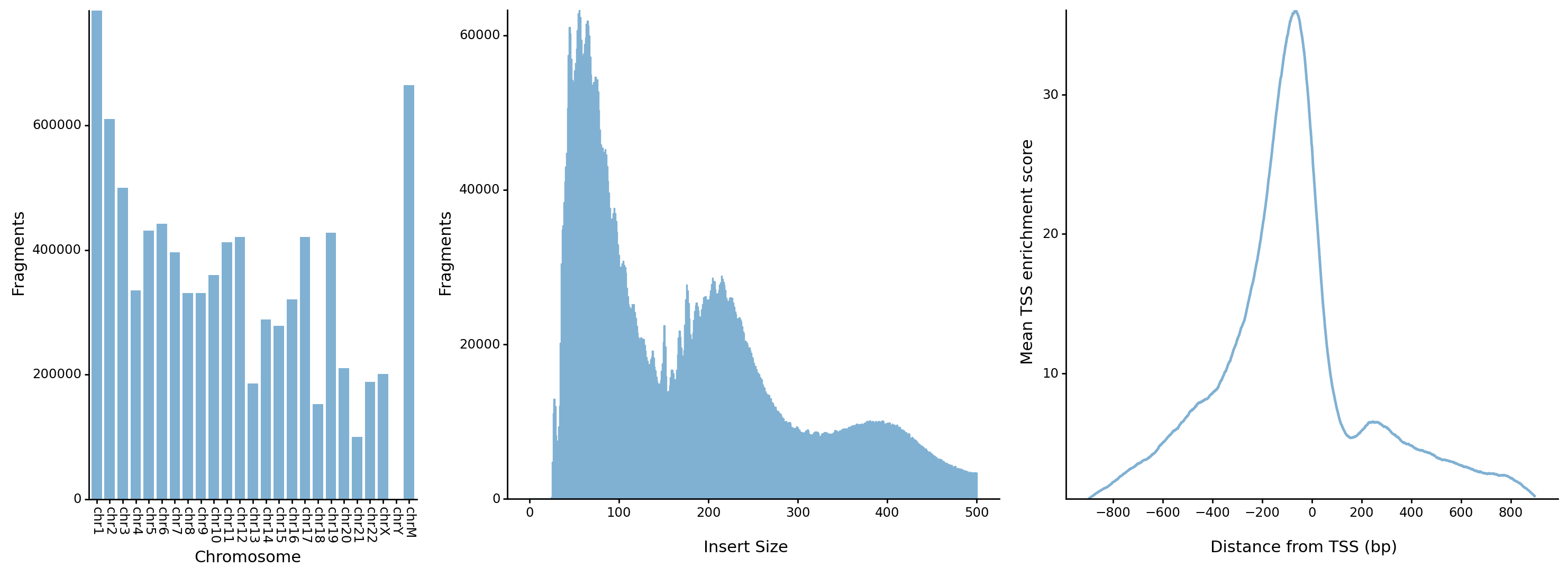
* The distrubution of fragments in chromosomes
* The distrubution of fragment lengths
* The distrubution of fragments around transcription start sites (TSSs)
* Other feature would be supported in the future ...
## Overview
%package -n python3-ATACFragQC
Summary: Fragment Quality Control for ATAC-seq
Provides: python-ATACFragQC
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-ATACFragQC
# ATACFragQC
The Python toolkits designed to control the fragment quality of Bulk/SingCell ATAC-seq.
## Installation
~~~
python3 -m pip install --upgrade ATACFragQC
~~~
## Usage
~~~
# Basic usage
ATACFragQC [options] -i <input.bam> -r <reference.gtf>
# For more information
ATACFragQC -h
~~~
## Features
* The distrubution of fragments in chromosomes
* The distrubution of fragment lengths
* The distrubution of fragments around transcription start sites (TSSs)
* Other feature would be supported in the future ...
## Overview

%package help
Summary: Development documents and examples for ATACFragQC
Provides: python3-ATACFragQC-doc
%description help
# ATACFragQC
The Python toolkits designed to control the fragment quality of Bulk/SingCell ATAC-seq.
## Installation
~~~
python3 -m pip install --upgrade ATACFragQC
~~~
## Usage
~~~
# Basic usage
ATACFragQC [options] -i <input.bam> -r <reference.gtf>
# For more information
ATACFragQC -h
~~~
## Features
* The distrubution of fragments in chromosomes
* The distrubution of fragment lengths
* The distrubution of fragments around transcription start sites (TSSs)
* Other feature would be supported in the future ...
## Overview

%prep
%autosetup -n ATACFragQC-0.4.6
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-ATACFragQC -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Tue May 30 2023 Python_Bot <Python_Bot@openeuler.org> - 0.4.6-1
- Package Spec generated
|
