1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
|
%global _empty_manifest_terminate_build 0
Name: python-TFBS-footprinting
Version: 1.0.0b54
Release: 1
Summary: Tool for identifying conserved TFBSs in vertebrate species.
License: MIT
URL: https://github.com/thirtysix/TFBS_footprinting
Source0: https://mirrors.nju.edu.cn/pypi/web/packages/46/7f/fb4affdfa2b2110330c718899a7ae8d143cb2c21882e1f00d4591bf868e2/TFBS_footprinting-1.0.0b54.tar.gz
BuildArch: noarch
Requires: python3-httplib2
Requires: python3-numpy
Requires: python3-matplotlib
Requires: python3-biopython
Requires: python3-msgpack
Requires: python3-wget
%description
## TFBS_footprinting
* This work is a derivative of ["Transcription factors"](https://commons.wikimedia.org/wiki/File:Transcription_Factors.svg) by [kelvin13](https://commons.wikimedia.org/wiki/User:Kelvin13), used under [CC BY 3.0](https://creativecommons.org/licenses/by/3.0/)
* * *
# Full documentation available at: [ReadTheDocs](https://tfbs-footprinting.readthedocs.io/en/latest/index.html)
## 1 Background
The TFBS footprinting method computationally predicts transcription factor binding sites (TFBSs) in a target species (e.g. homo sapiens) using 575 position weight matrices (PWMs) based on binding data from the JASPAR database. Additional experimental data from a variety of sources is used to support or detract from these predictions:
* DNA sequence conservation in homologous mammal species sequences
* proximity to CAGE-supported transcription start sites (TSSs)
* correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
* proximity to ChIP-Seq determined TFBSs (GTRD project)
* proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
* proximity to CpGs
* proximity to ATAC-Seq peaks (ENCODE project)
## 2 Output
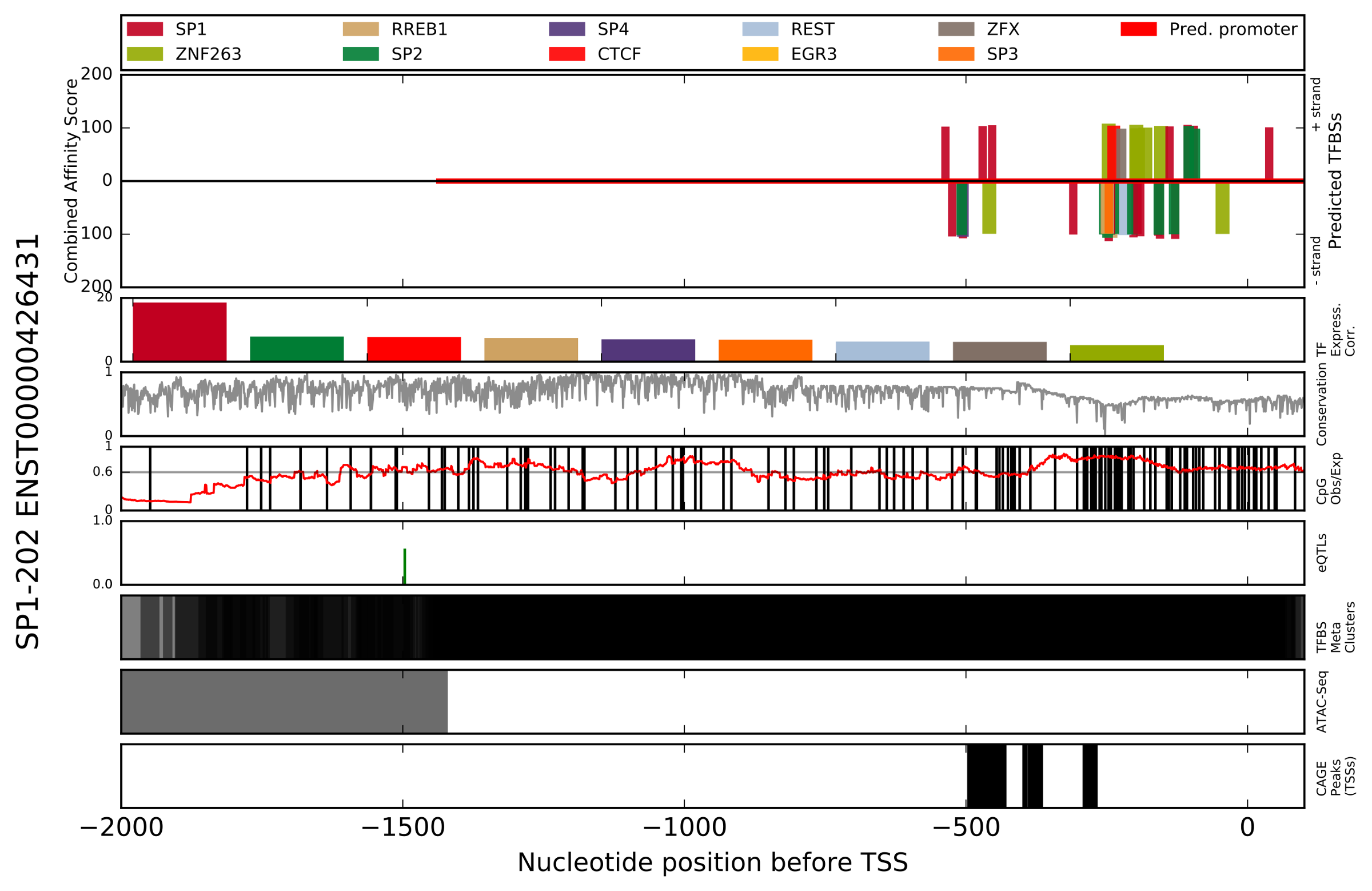
- Figure showing top_x_tfs highest scoring (combined affinity score) TFBSs mapped onto target_species promoter (ENSxxxxxxxxxxxx_[species_group].Promoterhisto.svg).
- Original alignment as retrieved from Ensembl (alignment_uncleaned.fasta).
- Cleaned alignment (alignment_cleaned.fasta).
- Regulatory information for the target transcripts user-defined promoter region (regulatory_decoded.json).
- Transcript properties for target transcript (transcript_dict.json).
- All predicted TFBSs for the target species which satisfy p-value threshold (TFBSs_found.all.json).
- All predicted TFBSs for target species which are supported by at least conservation_min predictions in other species, sorted by combined affinity score (TFBSs_found.sortedclusters.csv).
## 3 Installation
- Docker
`$ docker pull thirtysix/tfbs_footprinting`
- Pypi
`$ pip install tfbs_footprinting`
## 4 Usage
Predict TFBSs in the promoters any of 1-80,000 human protein coding transcripts in the Ensembl database. TFBS predictions can also be made for 87 unique non-human species (including model organisms such as mouse and zebrafish), present in the following groups:
- 70 Eutherian mammals
- 24 Primates
- 11 Fish
- 7 Sauropsids
View the available Ensembl species groups to plan your analysis: https://rest.ensembl.org/info/compara/species_sets/EPO_LOW_COVERAGE?content-type=application/json
### 4.1 Inputs
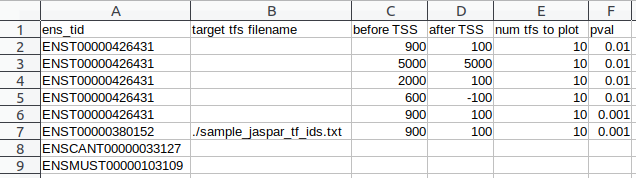
- Option 1: CSV of arguments
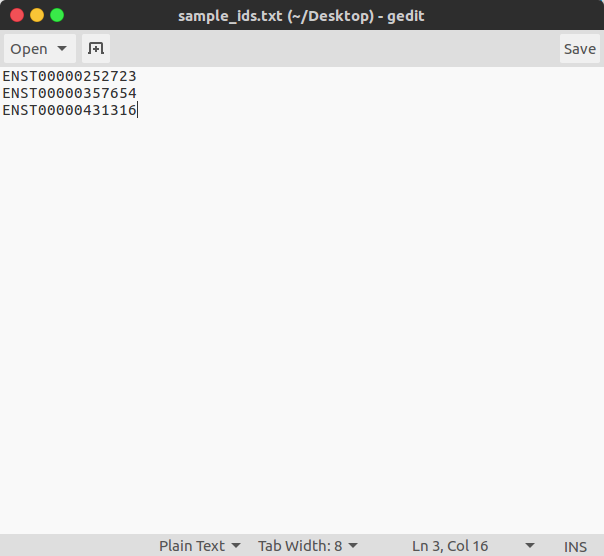
- Option 2: Simple text-file of Ensembl Transcript IDs
- File of Jaspar TF IDs (Not required)
### 4.2 TFBS_footprinter Use Examples
#### Running the sample analyses
- Run the sample analysis using a .csv of arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_analysis_list.csv`
- Run the sample analysis using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt`
#### Example using user-defined files/arguments
- Run the sample analysis using a .txt of Ensembl transcript ids, and all arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt -tfs PATH_TO/sample_analysis/sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -update`
#### Update the experimental data files
`$ tfbs_footprinter -update`
### 4.3 TFBS_footprinter Use Examples __(Within Docker)__
#### Running the sample analyses
1. Within Docker we first need to mount a volume so that the results of the analyis can be viewed on our host computer. It is recommended that you create an empty directory on your host computer:
`$ docker run -v /ABSOLUTE_PATH_TO/EMPTY_DIR_ON_HOST:/home/sample_analysis/tfbs_results -it thirtysix/tfbs_footprinting bash`
2. Then we move into the pre-existing sample analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./sample_analysis`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./sample_analysis_list.csv`
- Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$tfbs_footprinter -t ./sample_ensembl_ids.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./sample_ensembl_ids.txt -tfs ./sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o ./tfbs_results -update`
#### Example using user-defined files/arguments
1. Within Docker we first need to mount a volume so that we can __load your analysis files from your host computer__ into docker AND __save the results of the analysis on our host computer__:
`$ docker run -v /ABSOLUTE_PATH_TO/DIR_ON_HOST/CONTAINING_ANALYSIS_FILES:/home/analysis_dir -it thirtysix/tfbs_footprinting bash`
2. Then we move into your analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./analysis_dir`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./USER_TABLE_OF_ENSEMBL_IDS_AND_ARGS.csv`
3. Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt -tfs ./USER_LIST_OF_TF_NAMES.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o PATH_TO/Results/ -update`
### 4.4 Arguments
- --help, -h
show this help message and exit
- --t_ids_file, -t
Required for running an analysis. Location of a file containing Ensembl target_species transcript ids. Input options are either a text file of Ensembl transcript ids or a .csv file with individual values set for each parameter.
- --tf_ids_file, -tfs
Optional: Location of a file containing a limited list of Jaspar TFs to use in scoring alignment (see sample file tf_ids.txt at https://github.com/thirtysix/TFBS_footprinting)
[default: all Jaspar TFs]
- --target_species, -s
[default: "homo_sapiens"] - Target species (string), options are located at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species. Conservation of TFs across other species will be based on identifying them in this species first.
- --species_group, -g
("mammals", "primates", "sauropsids", or "fish")
[default: "mammals"] - Group of species (string) to identify conservation of TFs within. Your target species should be a member of this species group (e.g.
"homo_sapiens" and "mammals" or "primates"). The
"primates" group does not have a low-coverage version. Groups and members are listed at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species.
- --coverage, -e
("low" or "high") [default: "low"] - Which Ensembl EPO alignment of species to use. The low coverage contains significantly more species and is recommended. The primate group does not have a low-coverage version.
- --promoter_before_tss, -pb
(0-100,000) [default: 900] - Number (integer) of nucleotides upstream of TSS to include in analysis. If this number is negative the start point will be downstream of the TSS, the end point will then need to be further downstream.
- --promoter_after_tss, -pa
(0-100,000) [default: 100] - Number (integer) of nucleotides downstream of TSS to include in analysis. If this number is negative the end point will be upstream of the TSS. The start point will then need to be further upstream.
- --top_x_tfs, -tx
(1-20) [default: 10] - Number (integer) of unique TFs to include in output .svg figure.
- --pval, -p
P-value (float) for determine score cutoff (range: 0.1 to 0.0000001) [default: 0.01]
- --exp_data_update, -update
Download the latest experimental data files for use in analysis. Will run automatically if the "data" directory does not already exist (e.g. first usage).
## 5 Process
Iterate through each user provided Ensembl transcript id:
1. Retrieve EPO aligned orthologous sequences from Ensembl database for user-defined species group (mammals, primates, fish, sauropsids) for promoter of user-provided transcript id, between user-defined TSS-relative start/stop sites.
2. Edit retrieved alignment:
- Replace characters not corresponding to nucleotides (ACGT), with gaps characters "-".
- Remove gap-only columns from alignment.
3. Generate position weight matrices (PWMs) from Jaspar position frequency matrices (PFMs).
4. Score target species sequence using either all or a user-defined list of PWMs.
5. Keep predictions with a log-likelihood score greater than score threshold corresponding to p-value of 0.001, or user-defined p-value.
6. When experimental data is available for the target species, score each of the following for the target sequence region:
- DNA sequence conservation in homologous mammal species sequences
- proximity to CAGE-supported transcription start sites (TSSs)
- correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
- proximity to ChIP-Seq determined TFBSs (GTRD project)
- proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
- proximity to CpGs
- proximity to ATAC-Seq peaks (ENCODE project)
7. Compute 'combined affinity score' as a sum of scores for all experimental data.
8. Sort target_species predictions by combined affinity score, generate a vector graphics figure showing the top 10 (or user-defined) unique TFs mapped onto the promoter of the target transcript, and additional output as described below.
## 6 Species
The promoter region of any Ensembl transcript of any species within any column can be compared against the other members of the same column in order to identify a conserved binding site of the 575 transcription factors described in the Jaspar database. The Enredo-Pecan-Ortheus pipeline was used to create whole genome alignments between the species in each column. 'EPO_LOW' indicates this column also contains genomes for which the sequencing of the current version is still considered low-coverage. Due to the significantly greater number of species, we recommend using the low coverage versions except for primate comparisons which do not have a low coverage version. This list may not fully resp
|EPO_LOW mammals |EPO_LOW fish |EPO_LOW sauropsids |EPO mammals |EPO primates |EPO fish |EPO sauropsids |
|--------------------------|----------------------|-------------------|---------------------|-------------------|----------------------|-------------------|
|ailuropoda_melanoleuca |astyanax_mexicanus |anas_platyrhynchos |bos_taurus |callithrix_jacchus |danio_rerio |anolis_carolinensis|
|bos_taurus |danio_rerio |anolis_carolinensis|callithrix_jacchus |chlorocebus_sabaeus|gasterosteus_aculeatus|gallus_gallus |
|callithrix_jacchus |gadus_morhua |ficedula_albicollis|canis_familiaris |gorilla_gorilla |lepisosteus_oculatus |meleagris_gallopavo|
|canis_familiaris |gasterosteus_aculeatus|gallus_gallus |chlorocebus_sabaeus |homo_sapiens |oryzias_latipes |taeniopygia_guttata|
|cavia_porcellus |lepisosteus_oculatus |meleagris_gallopavo|equus_caballus |macaca_mulatta |tetraodon_nigroviridis| |
|chlorocebus_sabaeus |oreochromis_niloticus |pelodiscus_sinensis|felis_catus |pan_troglodytes | | |
|choloepus_hoffmanni |oryzias_latipes |taeniopygia_guttata|gorilla_gorilla |papio_anubis | | |
|dasypus_novemcinctus |poecilia_formosa | |homo_sapiens |pongo_abelii | | |
|dipodomys_ordii |takifugu_rubripes | |macaca_mulatta | | | |
|echinops_telfairi |tetraodon_nigroviridis| |mus_musculus | | | |
|equus_caballus |xiphophorus_maculatus | |oryctolagus_cuniculus| | | |
|erinaceus_europaeus | | |ovis_aries | | | |
|felis_catus | | |pan_troglodytes | | | |
|gorilla_gorilla | | |papio_anubis | | | |
|homo_sapiens | | |pongo_abelii | | | |
|ictidomys_tridecemlineatus| | |rattus_norvegicus | | | |
|loxodonta_africana | | |sus_scrofa | | | |
|macaca_mulatta | | | | | | |
|microcebus_murinus | | | | | | |
|mus_musculus | | | | | | |
|mustela_putorius_furo | | | | | | |
|myotis_lucifugus | | | | | | |
|nomascus_leucogenys | | | | | | |
|ochotona_princeps | | | | | | |
|oryctolagus_cuniculus | | | | | | |
|otolemur_garnettii | | | | | | |
|ovis_aries | | | | | | |
|pan_troglodytes | | | | | | |
|papio_anubis | | | | | | |
|pongo_abelii | | | | | | |
|procavia_capensis | | | | | | |
|pteropus_vampyrus | | | | | | |
|rattus_norvegicus | | | | | | |
|sorex_araneus | | | | | | |
|sus_scrofa | | | | | | |
|tarsius_syrichta | | | | | | |
|tupaia_belangeri | | | | | | |
|tursiops_truncatus | | | | | | |
|vicugna_pacos | | | | | | |
%package -n python3-TFBS-footprinting
Summary: Tool for identifying conserved TFBSs in vertebrate species.
Provides: python-TFBS-footprinting
BuildRequires: python3-devel
BuildRequires: python3-setuptools
BuildRequires: python3-pip
%description -n python3-TFBS-footprinting
## TFBS_footprinting

* This work is a derivative of ["Transcription factors"](https://commons.wikimedia.org/wiki/File:Transcription_Factors.svg) by [kelvin13](https://commons.wikimedia.org/wiki/User:Kelvin13), used under [CC BY 3.0](https://creativecommons.org/licenses/by/3.0/)
* * *
# Full documentation available at: [ReadTheDocs](https://tfbs-footprinting.readthedocs.io/en/latest/index.html)
## 1 Background
The TFBS footprinting method computationally predicts transcription factor binding sites (TFBSs) in a target species (e.g. homo sapiens) using 575 position weight matrices (PWMs) based on binding data from the JASPAR database. Additional experimental data from a variety of sources is used to support or detract from these predictions:
* DNA sequence conservation in homologous mammal species sequences
* proximity to CAGE-supported transcription start sites (TSSs)
* correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
* proximity to ChIP-Seq determined TFBSs (GTRD project)
* proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
* proximity to CpGs
* proximity to ATAC-Seq peaks (ENCODE project)
## 2 Output
- Figure showing top_x_tfs highest scoring (combined affinity score) TFBSs mapped onto target_species promoter (ENSxxxxxxxxxxxx_[species_group].Promoterhisto.svg).

- Original alignment as retrieved from Ensembl (alignment_uncleaned.fasta).
- Cleaned alignment (alignment_cleaned.fasta).
- Regulatory information for the target transcripts user-defined promoter region (regulatory_decoded.json).
- Transcript properties for target transcript (transcript_dict.json).
- All predicted TFBSs for the target species which satisfy p-value threshold (TFBSs_found.all.json).
- All predicted TFBSs for target species which are supported by at least conservation_min predictions in other species, sorted by combined affinity score (TFBSs_found.sortedclusters.csv).
## 3 Installation
- Docker
`$ docker pull thirtysix/tfbs_footprinting`
- Pypi
`$ pip install tfbs_footprinting`
## 4 Usage
Predict TFBSs in the promoters any of 1-80,000 human protein coding transcripts in the Ensembl database. TFBS predictions can also be made for 87 unique non-human species (including model organisms such as mouse and zebrafish), present in the following groups:
- 70 Eutherian mammals
- 24 Primates
- 11 Fish
- 7 Sauropsids
View the available Ensembl species groups to plan your analysis: https://rest.ensembl.org/info/compara/species_sets/EPO_LOW_COVERAGE?content-type=application/json
### 4.1 Inputs
- Option 1: CSV of arguments

- Option 2: Simple text-file of Ensembl Transcript IDs

- File of Jaspar TF IDs (Not required)

### 4.2 TFBS_footprinter Use Examples
#### Running the sample analyses
- Run the sample analysis using a .csv of arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_analysis_list.csv`
- Run the sample analysis using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt`
#### Example using user-defined files/arguments
- Run the sample analysis using a .txt of Ensembl transcript ids, and all arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt -tfs PATH_TO/sample_analysis/sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -update`
#### Update the experimental data files
`$ tfbs_footprinter -update`
### 4.3 TFBS_footprinter Use Examples __(Within Docker)__
#### Running the sample analyses
1. Within Docker we first need to mount a volume so that the results of the analyis can be viewed on our host computer. It is recommended that you create an empty directory on your host computer:
`$ docker run -v /ABSOLUTE_PATH_TO/EMPTY_DIR_ON_HOST:/home/sample_analysis/tfbs_results -it thirtysix/tfbs_footprinting bash`
2. Then we move into the pre-existing sample analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./sample_analysis`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./sample_analysis_list.csv`
- Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$tfbs_footprinter -t ./sample_ensembl_ids.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./sample_ensembl_ids.txt -tfs ./sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o ./tfbs_results -update`
#### Example using user-defined files/arguments
1. Within Docker we first need to mount a volume so that we can __load your analysis files from your host computer__ into docker AND __save the results of the analysis on our host computer__:
`$ docker run -v /ABSOLUTE_PATH_TO/DIR_ON_HOST/CONTAINING_ANALYSIS_FILES:/home/analysis_dir -it thirtysix/tfbs_footprinting bash`
2. Then we move into your analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./analysis_dir`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./USER_TABLE_OF_ENSEMBL_IDS_AND_ARGS.csv`
3. Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt -tfs ./USER_LIST_OF_TF_NAMES.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o PATH_TO/Results/ -update`
### 4.4 Arguments
- --help, -h
show this help message and exit
- --t_ids_file, -t
Required for running an analysis. Location of a file containing Ensembl target_species transcript ids. Input options are either a text file of Ensembl transcript ids or a .csv file with individual values set for each parameter.
- --tf_ids_file, -tfs
Optional: Location of a file containing a limited list of Jaspar TFs to use in scoring alignment (see sample file tf_ids.txt at https://github.com/thirtysix/TFBS_footprinting)
[default: all Jaspar TFs]
- --target_species, -s
[default: "homo_sapiens"] - Target species (string), options are located at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species. Conservation of TFs across other species will be based on identifying them in this species first.
- --species_group, -g
("mammals", "primates", "sauropsids", or "fish")
[default: "mammals"] - Group of species (string) to identify conservation of TFs within. Your target species should be a member of this species group (e.g.
"homo_sapiens" and "mammals" or "primates"). The
"primates" group does not have a low-coverage version. Groups and members are listed at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species.
- --coverage, -e
("low" or "high") [default: "low"] - Which Ensembl EPO alignment of species to use. The low coverage contains significantly more species and is recommended. The primate group does not have a low-coverage version.
- --promoter_before_tss, -pb
(0-100,000) [default: 900] - Number (integer) of nucleotides upstream of TSS to include in analysis. If this number is negative the start point will be downstream of the TSS, the end point will then need to be further downstream.
- --promoter_after_tss, -pa
(0-100,000) [default: 100] - Number (integer) of nucleotides downstream of TSS to include in analysis. If this number is negative the end point will be upstream of the TSS. The start point will then need to be further upstream.
- --top_x_tfs, -tx
(1-20) [default: 10] - Number (integer) of unique TFs to include in output .svg figure.
- --pval, -p
P-value (float) for determine score cutoff (range: 0.1 to 0.0000001) [default: 0.01]
- --exp_data_update, -update
Download the latest experimental data files for use in analysis. Will run automatically if the "data" directory does not already exist (e.g. first usage).
## 5 Process
Iterate through each user provided Ensembl transcript id:
1. Retrieve EPO aligned orthologous sequences from Ensembl database for user-defined species group (mammals, primates, fish, sauropsids) for promoter of user-provided transcript id, between user-defined TSS-relative start/stop sites.
2. Edit retrieved alignment:
- Replace characters not corresponding to nucleotides (ACGT), with gaps characters "-".
- Remove gap-only columns from alignment.
3. Generate position weight matrices (PWMs) from Jaspar position frequency matrices (PFMs).
4. Score target species sequence using either all or a user-defined list of PWMs.
5. Keep predictions with a log-likelihood score greater than score threshold corresponding to p-value of 0.001, or user-defined p-value.
6. When experimental data is available for the target species, score each of the following for the target sequence region:
- DNA sequence conservation in homologous mammal species sequences
- proximity to CAGE-supported transcription start sites (TSSs)
- correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
- proximity to ChIP-Seq determined TFBSs (GTRD project)
- proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
- proximity to CpGs
- proximity to ATAC-Seq peaks (ENCODE project)
7. Compute 'combined affinity score' as a sum of scores for all experimental data.
8. Sort target_species predictions by combined affinity score, generate a vector graphics figure showing the top 10 (or user-defined) unique TFs mapped onto the promoter of the target transcript, and additional output as described below.
## 6 Species
The promoter region of any Ensembl transcript of any species within any column can be compared against the other members of the same column in order to identify a conserved binding site of the 575 transcription factors described in the Jaspar database. The Enredo-Pecan-Ortheus pipeline was used to create whole genome alignments between the species in each column. 'EPO_LOW' indicates this column also contains genomes for which the sequencing of the current version is still considered low-coverage. Due to the significantly greater number of species, we recommend using the low coverage versions except for primate comparisons which do not have a low coverage version. This list may not fully resp
|EPO_LOW mammals |EPO_LOW fish |EPO_LOW sauropsids |EPO mammals |EPO primates |EPO fish |EPO sauropsids |
|--------------------------|----------------------|-------------------|---------------------|-------------------|----------------------|-------------------|
|ailuropoda_melanoleuca |astyanax_mexicanus |anas_platyrhynchos |bos_taurus |callithrix_jacchus |danio_rerio |anolis_carolinensis|
|bos_taurus |danio_rerio |anolis_carolinensis|callithrix_jacchus |chlorocebus_sabaeus|gasterosteus_aculeatus|gallus_gallus |
|callithrix_jacchus |gadus_morhua |ficedula_albicollis|canis_familiaris |gorilla_gorilla |lepisosteus_oculatus |meleagris_gallopavo|
|canis_familiaris |gasterosteus_aculeatus|gallus_gallus |chlorocebus_sabaeus |homo_sapiens |oryzias_latipes |taeniopygia_guttata|
|cavia_porcellus |lepisosteus_oculatus |meleagris_gallopavo|equus_caballus |macaca_mulatta |tetraodon_nigroviridis| |
|chlorocebus_sabaeus |oreochromis_niloticus |pelodiscus_sinensis|felis_catus |pan_troglodytes | | |
|choloepus_hoffmanni |oryzias_latipes |taeniopygia_guttata|gorilla_gorilla |papio_anubis | | |
|dasypus_novemcinctus |poecilia_formosa | |homo_sapiens |pongo_abelii | | |
|dipodomys_ordii |takifugu_rubripes | |macaca_mulatta | | | |
|echinops_telfairi |tetraodon_nigroviridis| |mus_musculus | | | |
|equus_caballus |xiphophorus_maculatus | |oryctolagus_cuniculus| | | |
|erinaceus_europaeus | | |ovis_aries | | | |
|felis_catus | | |pan_troglodytes | | | |
|gorilla_gorilla | | |papio_anubis | | | |
|homo_sapiens | | |pongo_abelii | | | |
|ictidomys_tridecemlineatus| | |rattus_norvegicus | | | |
|loxodonta_africana | | |sus_scrofa | | | |
|macaca_mulatta | | | | | | |
|microcebus_murinus | | | | | | |
|mus_musculus | | | | | | |
|mustela_putorius_furo | | | | | | |
|myotis_lucifugus | | | | | | |
|nomascus_leucogenys | | | | | | |
|ochotona_princeps | | | | | | |
|oryctolagus_cuniculus | | | | | | |
|otolemur_garnettii | | | | | | |
|ovis_aries | | | | | | |
|pan_troglodytes | | | | | | |
|papio_anubis | | | | | | |
|pongo_abelii | | | | | | |
|procavia_capensis | | | | | | |
|pteropus_vampyrus | | | | | | |
|rattus_norvegicus | | | | | | |
|sorex_araneus | | | | | | |
|sus_scrofa | | | | | | |
|tarsius_syrichta | | | | | | |
|tupaia_belangeri | | | | | | |
|tursiops_truncatus | | | | | | |
|vicugna_pacos | | | | | | |
%package help
Summary: Development documents and examples for TFBS-footprinting
Provides: python3-TFBS-footprinting-doc
%description help
## TFBS_footprinting

* This work is a derivative of ["Transcription factors"](https://commons.wikimedia.org/wiki/File:Transcription_Factors.svg) by [kelvin13](https://commons.wikimedia.org/wiki/User:Kelvin13), used under [CC BY 3.0](https://creativecommons.org/licenses/by/3.0/)
* * *
# Full documentation available at: [ReadTheDocs](https://tfbs-footprinting.readthedocs.io/en/latest/index.html)
## 1 Background
The TFBS footprinting method computationally predicts transcription factor binding sites (TFBSs) in a target species (e.g. homo sapiens) using 575 position weight matrices (PWMs) based on binding data from the JASPAR database. Additional experimental data from a variety of sources is used to support or detract from these predictions:
* DNA sequence conservation in homologous mammal species sequences
* proximity to CAGE-supported transcription start sites (TSSs)
* correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
* proximity to ChIP-Seq determined TFBSs (GTRD project)
* proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
* proximity to CpGs
* proximity to ATAC-Seq peaks (ENCODE project)
## 2 Output
- Figure showing top_x_tfs highest scoring (combined affinity score) TFBSs mapped onto target_species promoter (ENSxxxxxxxxxxxx_[species_group].Promoterhisto.svg).

- Original alignment as retrieved from Ensembl (alignment_uncleaned.fasta).
- Cleaned alignment (alignment_cleaned.fasta).
- Regulatory information for the target transcripts user-defined promoter region (regulatory_decoded.json).
- Transcript properties for target transcript (transcript_dict.json).
- All predicted TFBSs for the target species which satisfy p-value threshold (TFBSs_found.all.json).
- All predicted TFBSs for target species which are supported by at least conservation_min predictions in other species, sorted by combined affinity score (TFBSs_found.sortedclusters.csv).
## 3 Installation
- Docker
`$ docker pull thirtysix/tfbs_footprinting`
- Pypi
`$ pip install tfbs_footprinting`
## 4 Usage
Predict TFBSs in the promoters any of 1-80,000 human protein coding transcripts in the Ensembl database. TFBS predictions can also be made for 87 unique non-human species (including model organisms such as mouse and zebrafish), present in the following groups:
- 70 Eutherian mammals
- 24 Primates
- 11 Fish
- 7 Sauropsids
View the available Ensembl species groups to plan your analysis: https://rest.ensembl.org/info/compara/species_sets/EPO_LOW_COVERAGE?content-type=application/json
### 4.1 Inputs
- Option 1: CSV of arguments

- Option 2: Simple text-file of Ensembl Transcript IDs

- File of Jaspar TF IDs (Not required)

### 4.2 TFBS_footprinter Use Examples
#### Running the sample analyses
- Run the sample analysis using a .csv of arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_analysis_list.csv`
- Run the sample analysis using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt`
#### Example using user-defined files/arguments
- Run the sample analysis using a .txt of Ensembl transcript ids, and all arguments:
`$ tfbs_footprinter -t PATH_TO/sample_analysis/sample_ensembl_ids.txt -tfs PATH_TO/sample_analysis/sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -update`
#### Update the experimental data files
`$ tfbs_footprinter -update`
### 4.3 TFBS_footprinter Use Examples __(Within Docker)__
#### Running the sample analyses
1. Within Docker we first need to mount a volume so that the results of the analyis can be viewed on our host computer. It is recommended that you create an empty directory on your host computer:
`$ docker run -v /ABSOLUTE_PATH_TO/EMPTY_DIR_ON_HOST:/home/sample_analysis/tfbs_results -it thirtysix/tfbs_footprinting bash`
2. Then we move into the pre-existing sample analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./sample_analysis`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./sample_analysis_list.csv`
- Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$tfbs_footprinter -t ./sample_ensembl_ids.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./sample_ensembl_ids.txt -tfs ./sample_jaspar_tf_ids.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o ./tfbs_results -update`
#### Example using user-defined files/arguments
1. Within Docker we first need to mount a volume so that we can __load your analysis files from your host computer__ into docker AND __save the results of the analysis on our host computer__:
`$ docker run -v /ABSOLUTE_PATH_TO/DIR_ON_HOST/CONTAINING_ANALYSIS_FILES:/home/analysis_dir -it thirtysix/tfbs_footprinting bash`
2. Then we move into your analysis directory in the Docker container to perform the analysis there so that the results generated there will automatically appear in the designated location on our host computer:
`$ cd ./analysis_dir`
3. Then we can run the sample analysis in Docker in the same way that we would normally use tfbs_footprinter (above), e.g. using a .csv of arguments:
`$ tfbs_footprinter -t ./USER_TABLE_OF_ENSEMBL_IDS_AND_ARGS.csv`
3. Or (again, as above) using a .txt of Ensembl transcript ids, and minimal arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt`
- Or (again, as above) using a .txt of Ensembl transcript ids, and multiple arguments:
`$ tfbs_footprinter -t ./USER_LIST_OF_ENSEMBL_IDS.txt -tfs ./USER_LIST_OF_TF_NAMES.txt -s homo_sapiens -g mammals -e low -pb 900 -pa 100 -tx 10 -o PATH_TO/Results/ -update`
### 4.4 Arguments
- --help, -h
show this help message and exit
- --t_ids_file, -t
Required for running an analysis. Location of a file containing Ensembl target_species transcript ids. Input options are either a text file of Ensembl transcript ids or a .csv file with individual values set for each parameter.
- --tf_ids_file, -tfs
Optional: Location of a file containing a limited list of Jaspar TFs to use in scoring alignment (see sample file tf_ids.txt at https://github.com/thirtysix/TFBS_footprinting)
[default: all Jaspar TFs]
- --target_species, -s
[default: "homo_sapiens"] - Target species (string), options are located at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species. Conservation of TFs across other species will be based on identifying them in this species first.
- --species_group, -g
("mammals", "primates", "sauropsids", or "fish")
[default: "mammals"] - Group of species (string) to identify conservation of TFs within. Your target species should be a member of this species group (e.g.
"homo_sapiens" and "mammals" or "primates"). The
"primates" group does not have a low-coverage version. Groups and members are listed at https://github.com/thirtysix/TFBS_footprinting/blob/master/README.md#6-species.
- --coverage, -e
("low" or "high") [default: "low"] - Which Ensembl EPO alignment of species to use. The low coverage contains significantly more species and is recommended. The primate group does not have a low-coverage version.
- --promoter_before_tss, -pb
(0-100,000) [default: 900] - Number (integer) of nucleotides upstream of TSS to include in analysis. If this number is negative the start point will be downstream of the TSS, the end point will then need to be further downstream.
- --promoter_after_tss, -pa
(0-100,000) [default: 100] - Number (integer) of nucleotides downstream of TSS to include in analysis. If this number is negative the end point will be upstream of the TSS. The start point will then need to be further upstream.
- --top_x_tfs, -tx
(1-20) [default: 10] - Number (integer) of unique TFs to include in output .svg figure.
- --pval, -p
P-value (float) for determine score cutoff (range: 0.1 to 0.0000001) [default: 0.01]
- --exp_data_update, -update
Download the latest experimental data files for use in analysis. Will run automatically if the "data" directory does not already exist (e.g. first usage).
## 5 Process
Iterate through each user provided Ensembl transcript id:
1. Retrieve EPO aligned orthologous sequences from Ensembl database for user-defined species group (mammals, primates, fish, sauropsids) for promoter of user-provided transcript id, between user-defined TSS-relative start/stop sites.
2. Edit retrieved alignment:
- Replace characters not corresponding to nucleotides (ACGT), with gaps characters "-".
- Remove gap-only columns from alignment.
3. Generate position weight matrices (PWMs) from Jaspar position frequency matrices (PFMs).
4. Score target species sequence using either all or a user-defined list of PWMs.
5. Keep predictions with a log-likelihood score greater than score threshold corresponding to p-value of 0.001, or user-defined p-value.
6. When experimental data is available for the target species, score each of the following for the target sequence region:
- DNA sequence conservation in homologous mammal species sequences
- proximity to CAGE-supported transcription start sites (TSSs)
- correlation of expression between target gene and predicted transcription factor (TF) across 1800+ samples
- proximity to ChIP-Seq determined TFBSs (GTRD project)
- proximity to qualitative trait loci (eQTLs) affecting expression of the target gene (GTEX project)
- proximity to CpGs
- proximity to ATAC-Seq peaks (ENCODE project)
7. Compute 'combined affinity score' as a sum of scores for all experimental data.
8. Sort target_species predictions by combined affinity score, generate a vector graphics figure showing the top 10 (or user-defined) unique TFs mapped onto the promoter of the target transcript, and additional output as described below.
## 6 Species
The promoter region of any Ensembl transcript of any species within any column can be compared against the other members of the same column in order to identify a conserved binding site of the 575 transcription factors described in the Jaspar database. The Enredo-Pecan-Ortheus pipeline was used to create whole genome alignments between the species in each column. 'EPO_LOW' indicates this column also contains genomes for which the sequencing of the current version is still considered low-coverage. Due to the significantly greater number of species, we recommend using the low coverage versions except for primate comparisons which do not have a low coverage version. This list may not fully resp
|EPO_LOW mammals |EPO_LOW fish |EPO_LOW sauropsids |EPO mammals |EPO primates |EPO fish |EPO sauropsids |
|--------------------------|----------------------|-------------------|---------------------|-------------------|----------------------|-------------------|
|ailuropoda_melanoleuca |astyanax_mexicanus |anas_platyrhynchos |bos_taurus |callithrix_jacchus |danio_rerio |anolis_carolinensis|
|bos_taurus |danio_rerio |anolis_carolinensis|callithrix_jacchus |chlorocebus_sabaeus|gasterosteus_aculeatus|gallus_gallus |
|callithrix_jacchus |gadus_morhua |ficedula_albicollis|canis_familiaris |gorilla_gorilla |lepisosteus_oculatus |meleagris_gallopavo|
|canis_familiaris |gasterosteus_aculeatus|gallus_gallus |chlorocebus_sabaeus |homo_sapiens |oryzias_latipes |taeniopygia_guttata|
|cavia_porcellus |lepisosteus_oculatus |meleagris_gallopavo|equus_caballus |macaca_mulatta |tetraodon_nigroviridis| |
|chlorocebus_sabaeus |oreochromis_niloticus |pelodiscus_sinensis|felis_catus |pan_troglodytes | | |
|choloepus_hoffmanni |oryzias_latipes |taeniopygia_guttata|gorilla_gorilla |papio_anubis | | |
|dasypus_novemcinctus |poecilia_formosa | |homo_sapiens |pongo_abelii | | |
|dipodomys_ordii |takifugu_rubripes | |macaca_mulatta | | | |
|echinops_telfairi |tetraodon_nigroviridis| |mus_musculus | | | |
|equus_caballus |xiphophorus_maculatus | |oryctolagus_cuniculus| | | |
|erinaceus_europaeus | | |ovis_aries | | | |
|felis_catus | | |pan_troglodytes | | | |
|gorilla_gorilla | | |papio_anubis | | | |
|homo_sapiens | | |pongo_abelii | | | |
|ictidomys_tridecemlineatus| | |rattus_norvegicus | | | |
|loxodonta_africana | | |sus_scrofa | | | |
|macaca_mulatta | | | | | | |
|microcebus_murinus | | | | | | |
|mus_musculus | | | | | | |
|mustela_putorius_furo | | | | | | |
|myotis_lucifugus | | | | | | |
|nomascus_leucogenys | | | | | | |
|ochotona_princeps | | | | | | |
|oryctolagus_cuniculus | | | | | | |
|otolemur_garnettii | | | | | | |
|ovis_aries | | | | | | |
|pan_troglodytes | | | | | | |
|papio_anubis | | | | | | |
|pongo_abelii | | | | | | |
|procavia_capensis | | | | | | |
|pteropus_vampyrus | | | | | | |
|rattus_norvegicus | | | | | | |
|sorex_araneus | | | | | | |
|sus_scrofa | | | | | | |
|tarsius_syrichta | | | | | | |
|tupaia_belangeri | | | | | | |
|tursiops_truncatus | | | | | | |
|vicugna_pacos | | | | | | |
%prep
%autosetup -n TFBS-footprinting-1.0.0b54
%build
%py3_build
%install
%py3_install
install -d -m755 %{buildroot}/%{_pkgdocdir}
if [ -d doc ]; then cp -arf doc %{buildroot}/%{_pkgdocdir}; fi
if [ -d docs ]; then cp -arf docs %{buildroot}/%{_pkgdocdir}; fi
if [ -d example ]; then cp -arf example %{buildroot}/%{_pkgdocdir}; fi
if [ -d examples ]; then cp -arf examples %{buildroot}/%{_pkgdocdir}; fi
pushd %{buildroot}
if [ -d usr/lib ]; then
find usr/lib -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/lib64 ]; then
find usr/lib64 -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/bin ]; then
find usr/bin -type f -printf "/%h/%f\n" >> filelist.lst
fi
if [ -d usr/sbin ]; then
find usr/sbin -type f -printf "/%h/%f\n" >> filelist.lst
fi
touch doclist.lst
if [ -d usr/share/man ]; then
find usr/share/man -type f -printf "/%h/%f.gz\n" >> doclist.lst
fi
popd
mv %{buildroot}/filelist.lst .
mv %{buildroot}/doclist.lst .
%files -n python3-TFBS-footprinting -f filelist.lst
%dir %{python3_sitelib}/*
%files help -f doclist.lst
%{_docdir}/*
%changelog
* Fri May 05 2023 Python_Bot <Python_Bot@openeuler.org> - 1.0.0b54-1
- Package Spec generated
|
